This repository contains the source code for the Monarch Initiative web application (aka, the Monarch UI).
This README describes the application and its interaction with Monarch Initiative services such as BioLink. It also provide a Quickstart on how to build and run the application.
For detailed information on the structure of the code and how to contribute, see CONTRIBUTING.md.
- Overall Architecture
- Directory Structure
- QuickStart
- Production Build: Bundling and Minification
- Production Test: Build and Serve
- Linting
- Testing
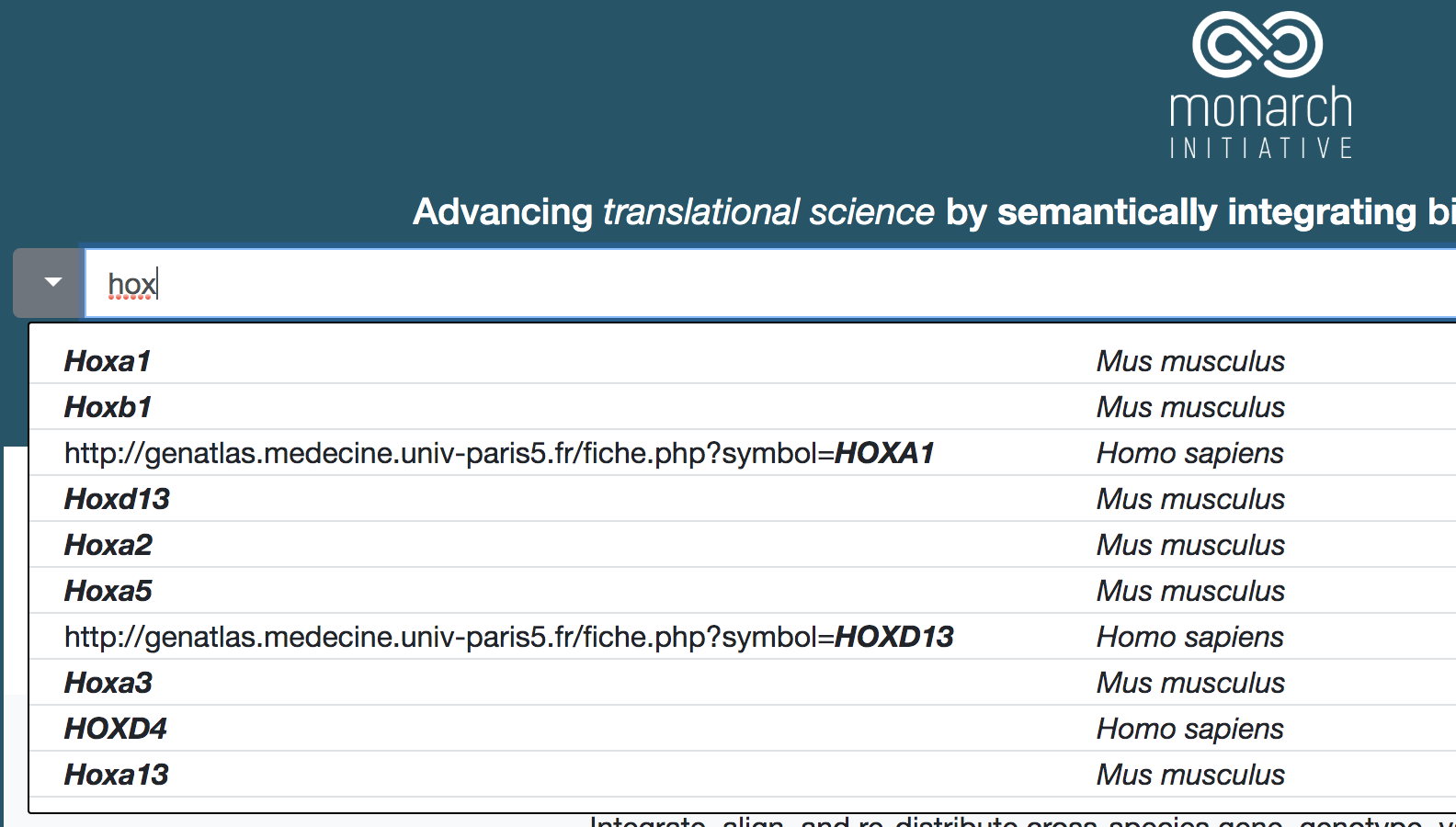
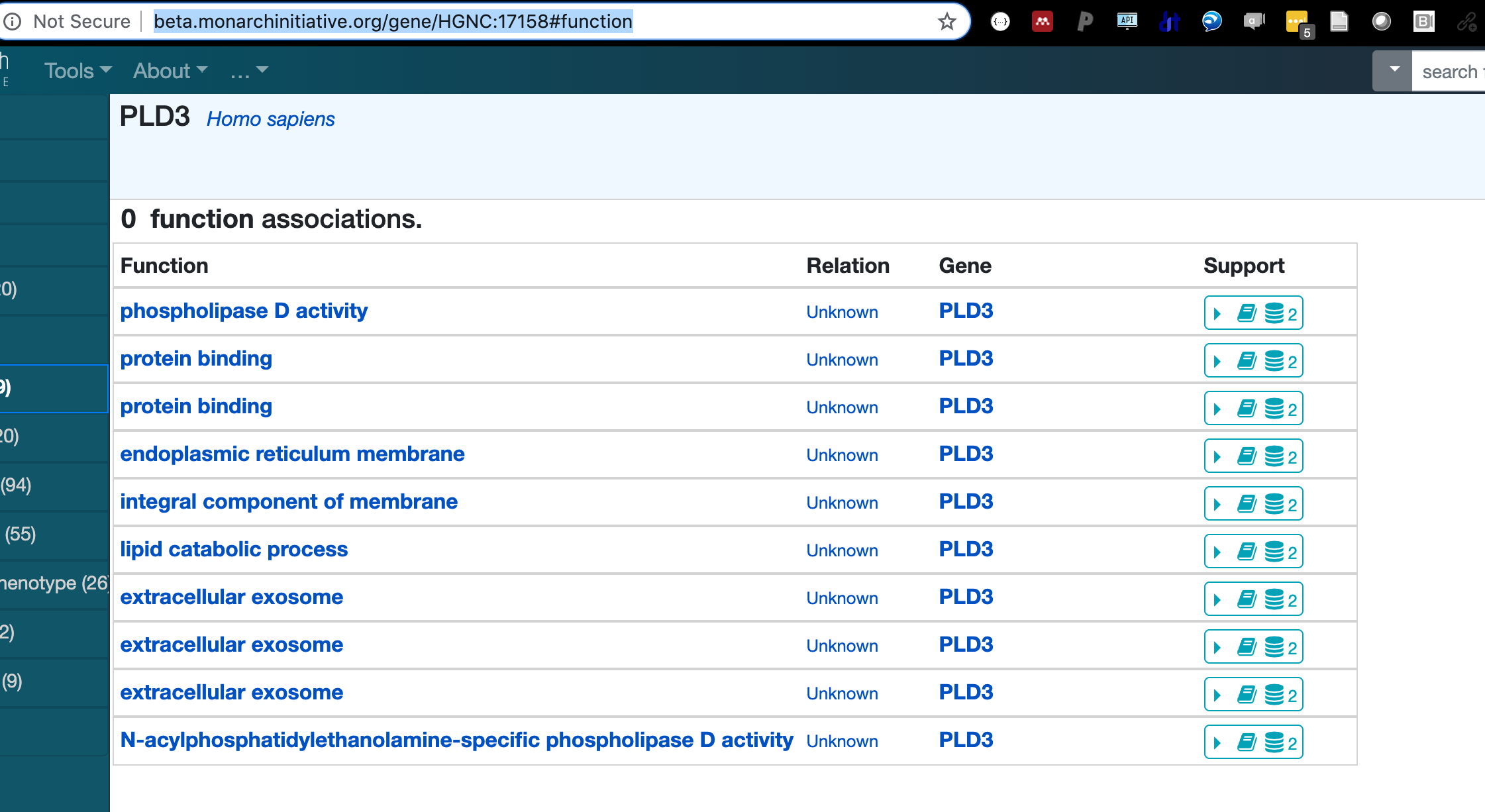
The UI is a VueJS single-page application that is loaded into the browser as a static set of Javascript, HTML, CSS, and media resources. Subsequent dynamic data is delivered to the browser via XMLHttpRequest calls to various backend services, primarily BioLink. The data returned from these calls is then displayed appropriately in the web application.
- Source: https://github.com/monarch-initiative/monarch-ui
- Monarch Initiative UI: https://monarchinitiative.org
- BioLink service to access Monarch's data
The VueJS application is built from source code via a modern chain of tools that deal with resource bundling, minification, and transpilation:
- VueJS UI Rendering framework
- BootstrapVue styling framework
- vue-cli for build and asset management
- webpack for asset bundling
The output of the build process is a set of static files including bundled Javascript, CSS, HTML and other media assets. These are then deployed to Netlify where they can be served to users on the internet.
There are multiple versions of the BioLink service.
See the top of the BioLink file in /api for what versions are available.
To easily switch between these versions in the live web-app, add a parameter to your current url like ?api=beta.
package.jsonlists build-time and run-time dependencies and a list ofnpm run-able scripts such asnpm run dev.vue.config.jsis the configuration that is used byvue-clito dynamically generate the configuration needed bywebpack, which is invoked behind the scenes byvue-cli.publish.shis a convenience script invoked bynpm run ghpublishto copy the developer's current directory into theirgh-pagesbranch and then pushing that toorigin.babel.config.jsconfigures the Babel compiler that translates modern Javascript into browser-compatible Javascript..eslintrc.jsspecifies the linting rules used by eslint to ensure that code is authored in a more uniform and safe way.
This directory contains the output of the build process (e.g., from npm run build or npm run ghpublish). The files here should never be placed in source control or edited; they are generated files. Upon first downloading the repo, or after npm run clean, this directory may not exist.
The vue-cli workflow specifies that the contents of public/ are copied verbatim to the build output directory dist/, and therefore made available to running web application.
team.yamldescribes the persons and institutions that make up the Monarch project. This is used byAboutTeam.mdto render a Team page.news.yamlis used to drive theHomeNews.mdcomponent.robots.txtis used to guide or exclude web-crawling robots. See Robots.txt.index.htmlis transformed by the build workflow to generate a correspondingdist/index.html. It is NOT copied verbatim.mondo_ids.txtis a tab-demimited list of "Diseases of the Month" for all 12 months of the year. It is used to populate the Featured Diseases carousel on the home page. The first field is the Mondo ID; the second indicates which month to display the disease, formatted as YYYY-MM-DD.
This directory and the monarch-ui/public/ (see above) directory contains the primary HTML, Javascript, and CSS source code that will be used to build the web application assets in monarch-ui/dist/.
By convention, vue-cli will use src/main.js as its main entry point. This file performs any global initialization, including the Vue router and the main App.vue.
By convention, App.vue is the root VueJS component that is usually bound to a div in the public/index.html file. This is a good place to configure VueJS and register global components.
This file router.js is where the vue-router is loaded and configured.
This file contains SCSS or CSS files that might need to be included in several components.
The most important file here is variables.scss, which can be included in every component via:
<style lang="scss">
@import "~@/style/variables";
...
</style>
We've tried to isolate external network API access into this directory, so that the rest of the UI can be unaware of the protocol and network issues inherent in making XMLHttpRequest or axios calls. For example, api/bio-link.js is the module that makes REST calls to the BioLink server, and the rest of the UI simply imports from api/bio-link.js. For example:
import * as BL from '@/api/bio-link';
// ...
const promise = BL.getNeighborhood(this.nodeId, this.nodeType);This directory is for non-UI utility code that does not access external network APIs (which would be located in src/api/).
Assets that will be bundled by webpack by require or import. E.g., require('../assets/img/icon-models.png')
Both components/ and views/ are properly VueJS components. However, by convention, we isolate top-level routable components into the views/ directory. Both component types are typically just VueJS Single-file Components with extension .vue. In some cases, they will have a .md extension, which means they will be scanned for Markdown prior to becoming VueJS components. See Markdown-based Components below for more information about Markdown in components.
In an attempt to make it easier for non-developers to contribute enhancements to the prose within the application, we've enabled the capability to define VueJS components that contain Markdown text in their <template>. These files with extension .md (e.g., views/AboutMonarch.md) are processed at build-time by loaders/vue-markdown-loader-improved.js, which will find Markdown fragments within the template and will translate them (at build-time) into HTML.
In order to build the application locally, you will need to obtain the source code and follow the QuickStart instructions below.
If your intent is to eventually contribute source code to the project via pull requests, then we recommend that you first fork the monarch-ui repo and then git clone this fork to your local development machine. Details on using the GitHub Flow workflow for contributing to open source projects are provided in CONTRIBUTING
For example, if your GitHub username is abc, and you've forked the monarch-ui repo to your user, then you can get a local copy of your forked repo with (assuming that monarch_stuff exists on your machine)
cd monarch_stuff/
git clone [email protected]:abc/monarch-ui.git
cd monarch_ui/
For simple evaluation or testing of the monarch-ui, then cloning the primary repo at monarchinitiative/monarch-ui should be sufficient. You will not be able to submit pull requests if you take this route, however.
cd monarch_stuff/
git clone https://github.com/monarch-initiative/monarch-ui.git
cd monarch_ui/
There may be some one-time installation of tools, depending upon whether you have the supported version of NodeJS running. NodeJS v8.12.0 is currently supported, although the project will likely build fine with later versions of NodeJS. If the following command fails, then you probably do not have NodeJS installed:
node -vIf the above command fails or if it reports a version number significantly different from v8.12.0, then you should install nvm - Node Version Manager and then install a compatible version of NodeJS.
In the event that you do not already have a compatible version of NodeJS installed, we have provided detailed instructions at the end of the CONTRIBUTING document here Installing NodeJS.
Whenever you update your local development machine's source code, and upon initially cloning the repository, you will need to run the following npm install command, which will read the latest downloaded package.json and ensure that your local dependencies are up to date.
npm install
The following command will run webpack in development mode, which runs a webpack-dev-server that detects source code changes and rebuilds the application dynamically and delivers these changes to the browser via hot reload, which often allows the UI to be updated without a full page reload. This speeds up a developer's iterations immensely.
npm run serve
The command npm run dev is an alias for npm run serve, mostly for the convenience of folks who are used to npm run dev.
In contrast to npm run serve above, which is designed to quickly build and demonstrate changes to the source code, the npm run build command will be more exhaustive in optimizing the build, and will produce a set of assets (Javascript, HTML, CSS, media) that are suitable for deployment to a web server.
npm run build
Although there shouldn't be any difference in behavior between development and production mode builds, there sometimes are. So it has proven convenient to be able to test the production build locally, which is why the npm run buildandserve command was added:
npm run buildandserve
Usually, we are editing code with a text editor (e.g., SublimeText) that works with ESLint and highlights any linting errors discovered as we edit. It is also possible to manually run ESLint over the entire source. This is a good idea to do before finalizing your code submission.
npm run lint
npm run test:unit
npm run test:e2e
npm run test