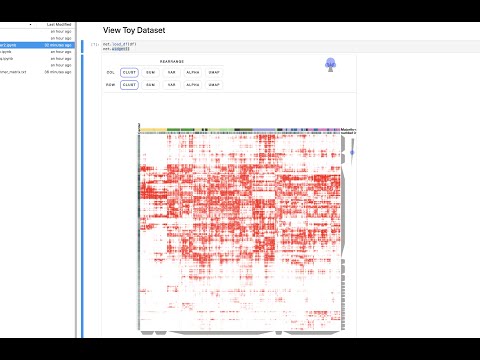
Clustergrammer is a web-based tool for visualizing high-dimensional data (e.g. a matrix) as an interactive and shareable hierarchically clustered heatmap. Clustergrammer's front end (Clustergrammer-JS) is built using D3.js and its back-end (Clustergrammer-PY) is built using Python. Clustergrammer produces highly interactive visualizations that enable intuitive exploration of high-dimensional data and has several biology-specific features (e.g. enrichment analysis, see Biology-Specific Features) to facilitate the exploration of gene-level biological data. Click the screenshot below to view an interactive tutorial:
Clustergrammer's interacive features include:
- Zooming and Panning
- Row and Column Reordering
- Interactive Dendrogram
- Interactive Dimensionality Reduction
- Interactive Categories
- Cropping
- Row Searching
- Biology-Specific Features
Clustergrammer can be used in three main ways (this repo contains the source code for Clustergrammer-JS):
- Clustergrammer Web App (http://amp.pharm.mssm.edu/clustergrammer/)
- Clustergrammer Jupyter Widget
- Clustergrammer-JS and Clustergrammer-PY libraries
For information about building a webpage or app using Clustergrammer see: Web-Development with Clustergrammer
Clustergrammer is being re-built using the WebGL library regl. The new in-development front-end is Clustergrammer-GL and the new in-development Jupyter widget is Clustergrammer2. The above notebook shows how Clustergrammer2 can be used to load a small dataset and visualize a large random DataFrame. By running the notebook on MyBinder using Jupyter Lab it can also be used to visualize a user uploaded dataset. Please see the video tutorial above for more information.
For additional examples and tutorials please see:
- Case Studies and Tutorials
- Clustergrammer2-Notebooks GitHub repository
Clustergrammer was recently presented at JupyterCon 2018 (see slides).
Pleae see Clustergramer's documentation for detailed information or select a specific topic below:
- Getting Started
- Interacting with the Visualization
- Web-Development with Clustergrammer (example pages)
- Clustergrammer Web App and Clustergrammer Jupyter Widget
- Matrix Formats and Input/Output
- Core libraries: Clustergrammer-JS and Clustergrammer-PY
- App Integration Examples
- Case Studies and Examples
- Biology-Specific Features
- Developing Clustergrammer
Please consider supporting Clustergrammer by citing our publication:
Fernandez, N. F. et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci. Data 4:170151 doi: 10.1038/sdata.2017.151 (2017).
Clustergrammer was developed by the Ma'ayan lab at the Icahn School of Medicine at Mount Sinai for the BD2K-LINCS DCIC and the KMC-IDG. Clustergrammer's license and third-party licenses are in the LICENSES directory and more information can be found at Clustergrammer License.
Please contact us for support, licensing questions, comments, and suggestions.