![[Oxford Nanopore Technologies]](/ONT_logo_deprecate.png)
We have new bioinformatic resources that replace the functionality of this project! For production modified base calling see the Dorado repository. For modified base data preparation, model training and experimental model testing see the Remora repository. For post-processing and analysis of modified base calling results see the modkit repository.
This repository is now unsupported and we do not recommend its use. Please contact Oxford Nanopore: [email protected] for help with your application if it is not possible to upgrade.
Megalodon is a research command line tool to extract high accuracy modified base and sequence variant calls from raw nanopore reads by anchoring the information rich basecalling neural network output to a reference genome/transcriptome.
Raw nanopore reads are processed by a single command to produce basecalls (FASTA/Q), reference mappings (SAM/BAM/CRAM), modified base calls (per-read and bedgraph/bedmethyl/modVCF), sequence variant calls (per-read and VCF) and more.
Detailed documentation for all megalodon commands and algorithms can be found on the Megalodon documentation page.
The primary Megalodon run mode requires the Guppy basecaller (version >= 4.0). See the community page for download/installation instructions [login required].
Megalodon is a python-based command line software package.
Given a python (version >= 3.6) installation, all other requirements are handled by pip or conda.
Taiyaki is no longer required to run Megalodon, but installation is required for two specific run modes:
- output mapped signal files (for basecall model training)
- running the Taiyaki basecalling backend (for neural network designs including experimental layers)
The ont-pyguppy-client-api dependency provides select release specifications, so compatible python versions and operating systems may be limited.
pip is recommended for Megalodon installation.
pip install megalodon
conda installation is available, but not fully supported.
ont_pyguppy_client_lib is not available on conda and thus must be installed with pip.
conda install megalodon pip install ont_pyguppy_client_lib
To install from github source for development, the following commands can be run.
git clone https://github.com/nanoporetech/megalodon pip install -e megalodon/
Megalodon must obtain the intermediate output from the basecall neural network.
Guppy (production nanopore basecalling software) is the recommended backend to obtain this output from raw nanopore signal (from FAST5 files).
Nanopore basecalling is compute intensive and thus it is highly recommended that GPU resources are specified (--devices) for optimal Megalodon performance.
Megalodon is accessed via the command line interface megalodon command.
# megalodon help (common args)
megalodon -h
# megalodon help (all args)
megalodon --help-long
# Example command to output basecalls, mappings, and CpG 5mC and 5hmC methylation in both per-read (``mod_mappings``) and aggregated (``mods``) formats
# Compute settings: GPU devices 0 and 1 with 20 CPU cores
megalodon \
raw_fast5s/ \
--guppy-config dna_r9.4.1_450bps_fast.cfg \
--remora-modified-bases dna_r9.4.1_e8 fast 0.0.0 5hmc_5mc CG 0 \
--outputs basecalls mappings mod_mappings mods \
--reference reference.fa \
--devices 0 1 \
--processes 20
The above command uses the modified base model included in Remora. For more details on Remora modified base settings see the Remora repository.
This command produces the megalodon_results output directory containing all requested output files and logs.
The format for common outputs is described briefly below and in more detail in the full documentation
The code below shows how to obtain and run the R9.4.1, MinION/GridION, 5mC CpG model (same model shipped with Guppy as of 4.5.2 release).
# Obtain and run R9.4.1, MinION, 5mC CpG model from Rerio
git clone https://github.com/nanoporetech/rerio
rerio/download_model.py rerio/basecall_models/res_dna_r941_min_modbases_5mC_CpG_v001
megalodon \
raw_fast5s/ \
--guppy-params "-d ./rerio/basecall_models/" \
--guppy-config res_dna_r941_min_modbases_5mC_CpG_v001.cfg \
--outputs basecalls mappings mod_mappings mods \
--reference reference.fa \
--mod-motif m CG 0 \
--devices 0 1 \
--processes 20
The path to theguppy_basecall_serverexecutable is required to run Megalodon. By default, Megalodon assumes Guppy (Linux GPU) is installed in the current working directory (i.e../ont-guppy/bin/guppy_basecall_server). Use the--guppy-server-pathargument to specify a different path.
- Raw reads
- Directory containing raw read FAST5 files (sub-directories recursively searched)
- Reference
- Genome or transcriptome sequence reference (FASTA or minimap2 index)
- Variants File
- Megalodon requires a set of candidate variants for
--outputs variants(provide via--variant-filenameargument; VCF or BCF).
- Megalodon requires a set of candidate variants for
All Megalodon outputs are written into the directory specified with the --output-directory option with standard file names and extensions.
- Basecalls
- Format: FASTQ (default) or FASTA
- Basecall-anchored modified base scores are also available in hts-spec BAM format tags (
--outputs mod_basecalls).
- Mappings
- Format: SAM, BAM (default), or CRAM
- A tab-separated mapping text summary is also produced including per-read alignment statistics.
- Modified Base Calls
- The basecalling model specifies the modified bases capable of being output. See
megalodon_extras modified_bases describe_alphabet. - Per-read modified base calls
- SQL DB containing per-read modified base scores at each covered reference location
- Reference-anchored per-read modified base calls is BAM format via the
MmandMltags (see hts-spec specifications here).
- Aggregated calls
- Format: bedgraph, bedmethyl (default), and/or modVCF
- In order to restrict modified base calls to a specific motif(s) specify the
--mod-motifargument. For example, to restrict calls to CpG sites specify--mod-motif Z CG 0.
- The basecalling model specifies the modified bases capable of being output. See
- Sequence Variant Calls
- Per-read Variant Calls
- SQL DB containing per-read variant scores for each covered variant
- Aggregated calls
- Format: VCF
- Default run mode is diploid. To run in haploid mode, set
--haploidflag. - For best results on a diploid genome see the variant phasing workflow on the full documentation page.
- Per-read Variant Calls
Megalodon supports live run processing.
Activate live processing mode by simply adding the --live-processing argument and specifying the MinKNOW output directory as the Megalodon FAST5 input directory.
Megalodon will continue to search for FAST5s until the final_summary* file is created by MinKNOW, indicating data production has completed.
The basecalling model defines the modified bases capable of being output by Megalodon. Basecalling models must be trained to specifically detect a type or types of modified bases. See the Megalodon documentation here for instructions to construct modified base training data and train a new modified base model.
By default, Megalodon uses the dna_r9.4.1_450bps_modbases_5mc_hac.cfg Guppy config (released in version 4.5.2).
This config is compatible with DNA, R9.4.1, MinION/GridION reads and allows output of 5mC calls in all contexts.
Use the --guppy-config option to specify a different guppy model config.
The appropriate Rerio model is recommended for the highest accuracy modified base calls.
All configs can be used to output basecalls and mappings (as well as signal_mappings and per_read_refs for basecall training).
Modified base and sequence variant outputs require Megalodon calibration files.
To list configs with default calibration files, run megalodon --list-supported-guppy-configs.
See calibration documentation here for details on Megalodon model calibration.
Only flip-flop configs/models are currently supported by Megalodon (this excludes k-mer based and RLE model types).
In addition to the --guppy-config and --guppy-server-path options, a number of additional arguments control the behavior of the guppy backend.
The --guppy-params argument will pass arguments directly to the guppy_basecall_server initialization call.
For example to optimize GPU usage, the following option might be specified: --guppy-params "--num_callers 5 --ipc_threads 6"
Finally the --guppy-timeout arguments ensures that a run will not stall on few reads or with lower compute resources.
The Guppy server unable to recieve read error indicate that the Guppy server is overwhelmed.
Consider lowering the --processes and/or --guppy-concurrent-reads values to reduce these errors.
Finding the right balance for these parameters can help achieve optimal performance on a system.
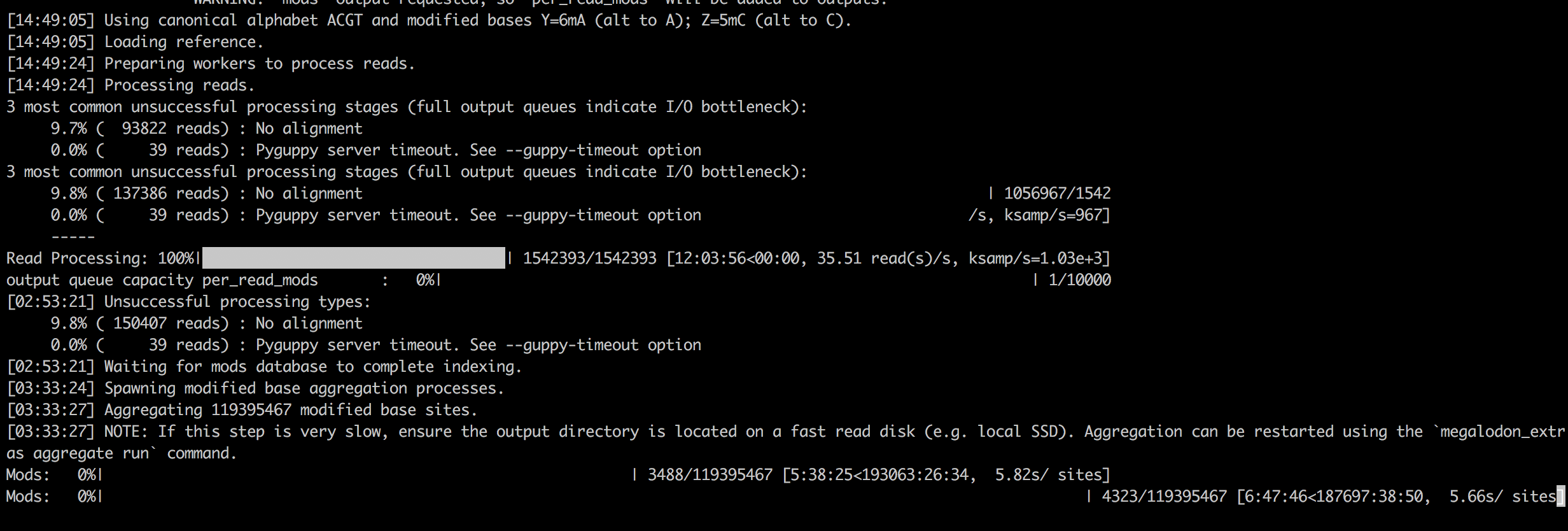
The status of the extract signal input queue and output queues is displayed by default (suppress with --suppress-queues-status).
If the extract_signal input queue is often empty, Megalodon is waiting on reading raw signal from FAST5 files.
If the input queue remains empty, increasing the --num-read-enumeration-threads and/or --num-extract-signal-processes parameters (defaults 8 and 2)) may improve performance.
Note that [--num-read-enumeration-threads] threads will be opened within each extract signal process.
Alternatively and if available, the input FAST5s disk location could be moved to faster I/O disk.
If any output status bars indicate a full queue, Megalodon will stall waiting on that process to write data to disk.
Moving the --output-directory accordingly to a location with faster disk I/O performance should improve performance.
Per-read modified base and variant statistics are stored in an on-disk sqlite database, which can be very dependent on disk speed and configuration.
In order to obtain the highest quality diploid sequence variant calls, the full variant phasing pipeline employing whatshap should be applied.
This pipeline is described in detail on the full documentation page.
The default diploid variant settings are optimized for the full phasing pipeline and not the highest quality diploid calls directly from a single Megalodon call.
When running Megalodon with a high density of variants (more than 1 variant per 100 reference bases), certain steps can be taken to increase performance. See variant atomize documentation for further details.
Megalodon supports processing direct RNA nanopore data.
In order to process an RNA sample specify the --rna flag as well as an RNA model using the --guppy-config argument.
Megalodon performs mapping using the standard minimap2 option, map-ont, and not the splice option, so a transcriptome reference must be provided.
The Megalodon code supports RNA modified base detection, but currently no RNA modified base basecalling models are released.
Megalodon does not currently perform checking that a set of reads agree with the provided model or options specified (e.g. --rna).
Users should take care to ensure that the correct options are specified for each sample processed.
© 2019-21 Oxford Nanopore Technologies Ltd.
Megalodon is distributed under the terms of the Oxford Nanopore Technologies, Ltd. Public License, v. 1.0. If a copy of the License was not distributed with this file, You can obtain one at http://nanoporetech.com
Research releases are provided as technology demonstrators to provide early access to features or stimulate Community development of tools. Support for this software will be minimal and is only provided directly by the developers. Feature requests, improvements, and discussions are welcome and can be implemented by forking and pull requests. However much as we would like to rectify every issue and piece of feedback users may have, the developers may have limited resource for support of this software. Research releases may be unstable and subject to rapid iteration by Oxford Nanopore Technologies.