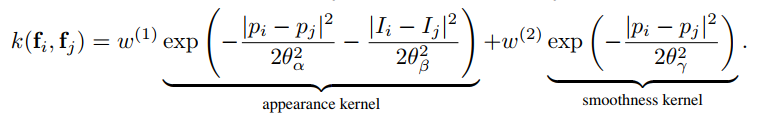This is a (Cython-based) Python wrapper for Philipp Krähenbühl's Fully-Connected CRFs (version 2, new, incomplete page).
If you use this code for your reasearch, please cite:
Efficient Inference in Fully Connected CRFs with Gaussian Edge Potentials
Philipp Krähenbühl and Vladlen Koltun
NIPS 2011
and provide a link to this repository as a footnote or a citation.
The package is on PyPI, so simply run pip install pydensecrf to install it.
If you want the newest and freshest version, you can install it by executing:
pip install git+https://github.com/lucasb-eyer/pydensecrf.git
and ignoring all the warnings coming from Eigen.
Note that you need a relatively recent version of Cython (at least version 0.22) for this wrapper,
the one shipped with Ubuntu 14.04 is too old. (Thanks to Scott Wehrwein for pointing this out.)
I suggest you use a virtual environment and install
the newest version of Cython there (pip install cython), but you may update the system version by
sudo apt-get remove cython
sudo pip install -U cython
Since this library needs to compile C++ code, installation can be a little more problematic than pure Python packages. Make sure to have Cython installed or try installing via conda instead if you are getting problems. PRs that improve Windows support are welcome.
pydensecrf does not come pre-installed in Colab or Kaggle Kernel. Running pip install pydensecrf will result into
build failures. Follow these steps instead for Colab/Kaggle Kernel:
pip install -U cython
pip install git+https://github.com/lucasb-eyer/pydensecrf.git
For images, the easiest way to use this library is using the DenseCRF2D class:
import numpy as np
import pydensecrf.densecrf as dcrf
d = dcrf.DenseCRF2D(640, 480, 5) # width, height, nlabelsYou can then set a fixed unary potential in the following way:
U = np.array(...) # Get the unary in some way.
print(U.shape) # -> (5, 480, 640)
print(U.dtype) # -> dtype('float32')
U = U.reshape((5,-1)) # Needs to be flat.
d.setUnaryEnergy(U)
# Or alternatively: d.setUnary(ConstUnary(U))Remember that U should be negative log-probabilities, so if you're using
probabilities py, don't forget to U = -np.log(py) them.
Requiring the reshape on the unary is an API wart that I'd like to fix, but
don't know how to without introducing an explicit dependency on numpy.
Note that the nlabels dimension is the first here before the reshape;
you may need to move it there before reshaping if that's not already the case,
like so:
print(U.shape) # -> (480, 640, 5)
U = U.transpose(2, 0, 1).reshape((5,-1))There's two common ways of getting unary potentials:
-
From a hard labeling generated by a human or some other processing. This case is covered by
from pydensecrf.utils import unary_from_labels. -
From a probability distribution computed by, e.g. the softmax output of a deep network. For this, see
from pydensecrf.utils import unary_from_softmax.
For usage of both of these, please refer to their docstrings or have a look at the example.
The two-dimensional case has two utility methods for adding the most-common pairwise potentials:
# This adds the color-independent term, features are the locations only.
d.addPairwiseGaussian(sxy=(3,3), compat=3, kernel=dcrf.DIAG_KERNEL, normalization=dcrf.NORMALIZE_SYMMETRIC)
# This adds the color-dependent term, i.e. features are (x,y,r,g,b).
# im is an image-array, e.g. im.dtype == np.uint8 and im.shape == (640,480,3)
d.addPairwiseBilateral(sxy=(80,80), srgb=(13,13,13), rgbim=im, compat=10, kernel=dcrf.DIAG_KERNEL, normalization=dcrf.NORMALIZE_SYMMETRIC)Both of these methods have shortcuts and default-arguments such that the most common use-case can be simplified to:
d.addPairwiseGaussian(sxy=3, compat=3)
d.addPairwiseBilateral(sxy=80, srgb=13, rgbim=im, compat=10)The parameters map to those in the paper as follows: sxy in the Gaussian case is $\theta_{\gamma}$,
and in the Bilateral case, sxy and srgb map to $\theta_{\alpha}$ and $\theta_{\beta}$, respectively.
The names are shorthand for "x/y standard-deviation" and "rgb standard-deviation" and for reference, the formula is:
An important caveat is that addPairwiseBilateral only works for RGB images, i.e. three channels.
If your data is of different type than this simple but common case, you'll need to compute your
own pairwise energy using utils.create_pairwise_bilateral; see the generic non-2D case for details.
A good example of working with Non-RGB data is provided as a notebook in the examples folder.
The compat argument can be any of the following:
- A number, then a
PottsCompatibilityis being used. - A 1D array, then a
DiagonalCompatibilityis being used. - A 2D array, then a
MatrixCompatibilityis being used.
These are label-compatibilites µ(xi, xj) whose parameters could possibly be learned.
For example, they could indicate that mistaking bird pixels for sky is not as bad as mistaking cat for sky.
The arrays should have nlabels or (nlabels,nlabels) as shape and a float32 datatype.
Possible values for the kernel argument are:
CONST_KERNELDIAG_KERNEL(the default)FULL_KERNEL
This specifies the kernel's precision-matrix Λ(m), which could possibly be learned.
These indicate correlations between feature types, the default implying no correlation.
Again, this could possiblty be learned.
Possible values for the normalization argument are:
NO_NORMALIZATIONNORMALIZE_BEFORENORMALIZE_AFTERNORMALIZE_SYMMETRIC(the default)
I have so far not found a way to set the kernel weights w(m).
According to the paper, w(2) was set to 1 and w(1) was cross-validated, but never specified.
Looking through Philip's code (included in pydensecrf/densecrf),
I couldn't find such explicit weights, and my guess is they are thus hard-coded to 1.
If anyone knows otherwise, please open an issue or, better yet, a pull-request.
Update: user @waldol1 has an idea in this issue. Feel free to try it out!
The easiest way to do inference with 5 iterations is to simply call:
Q = d.inference(5)And the MAP prediction is then:
map = np.argmax(Q, axis=0).reshape((640,480))If you're interested in the class-probabilities Q, you'll notice Q is a
wrapped Eigen matrix. The Eigen wrappers of this project implement the buffer
interface and can be simply cast to numpy arrays like so:
proba = np.array(Q)If for some reason you want to run the inference loop manually, you can do so:
Q, tmp1, tmp2 = d.startInference()
for i in range(5):
print("KL-divergence at {}: {}".format(i, d.klDivergence(Q)))
d.stepInference(Q, tmp1, tmp2)The DenseCRF class can be used for generic (non-2D) dense CRFs.
Its usage is exactly the same as above, except that the 2D-specific pairwise
potentials addPairwiseGaussian and addPairwiseBilateral are missing.
Instead, you need to use the generic addPairwiseEnergy method like this:
d = dcrf.DenseCRF(100, 5) # npoints, nlabels
feats = np.array(...) # Get the pairwise features from somewhere.
print(feats.shape) # -> (7, 100) = (feature dimensionality, npoints)
print(feats.dtype) # -> dtype('float32')
dcrf.addPairwiseEnergy(feats)In addition, you can pass compatibility, kernel and normalization
arguments just like in the 2D gaussian and bilateral cases.
The potential will be computed as w*exp(-0.5 * |f_i - f_j|^2).
User @markusnagel has written a couple numpy-functions generalizing the two
classic 2-D image pairwise potentials (gaussian and bilateral) to an arbitrary
number of dimensions: create_pairwise_gaussian and create_pairwise_bilateral.
You can access them as from pydensecrf.utils import create_pairwise_gaussian
and then have a look at their docstring to see how to use them.
The learning has not been fully wrapped. If you need it, get in touch or better yet, wrap it and submit a pull-request!
Here's a pointer for starters: issue#24. We need to wrap the gradients and getting/setting parameters. But then, we also need to do something with these, most likely call minimizeLBFGS from optimization.cpp. It should be relatively straightforward to just follow the learning examples included in the original code.
If while importing pydensecrf you get an error about some undefined symbols (for example .../pydensecrf/densecrf.so: undefined symbol: _ZTINSt8ios_base7failureB5cxx11E), you most likely are inadvertently mixing different compilers or toolchains. Try to see what's going on using tools like ldd. If you're using Anaconda, running conda install libgcc might be a solution.
This is a pretty common user error.
It means exactly what it says: you are passing a double but it wants a float.
Solve it by, for example, calling d.setUnaryEnergy(U.astype(np.float32)) instead of just d.setUnaryEnergy(U), or using float32 in your code in the first place.
My results are all pixelated like MS Paint's airbrush tool!
You screwed up reshaping somewhere and treated the class/label dimension as spatial dimension. This is you misunderstanding NumPy's memory layout and nothing that PyDenseCRF can detect or help with.
This mistake often happens for the Unary, see the Note in that section of the README.
These are instructions for maintainers about how to release new versions. (Mainly instructions for myself.)
# Go increment the version in setup.py
> python setup.py build_ext
> python setup.py sdist
> twine upload dist/pydensecrf-VERSION_NUM.tar.gz
And that's it. At some point, it would be cool to automate this on TravisCI, but not worth it yet. At that point, looking into creating "manylinux" wheels might be nice, too.
Thanks to @MarvinTeichmann we now have proper tests, install the package and run py.test.
Maybe there's a better way to run them, but both of us don't know 😄