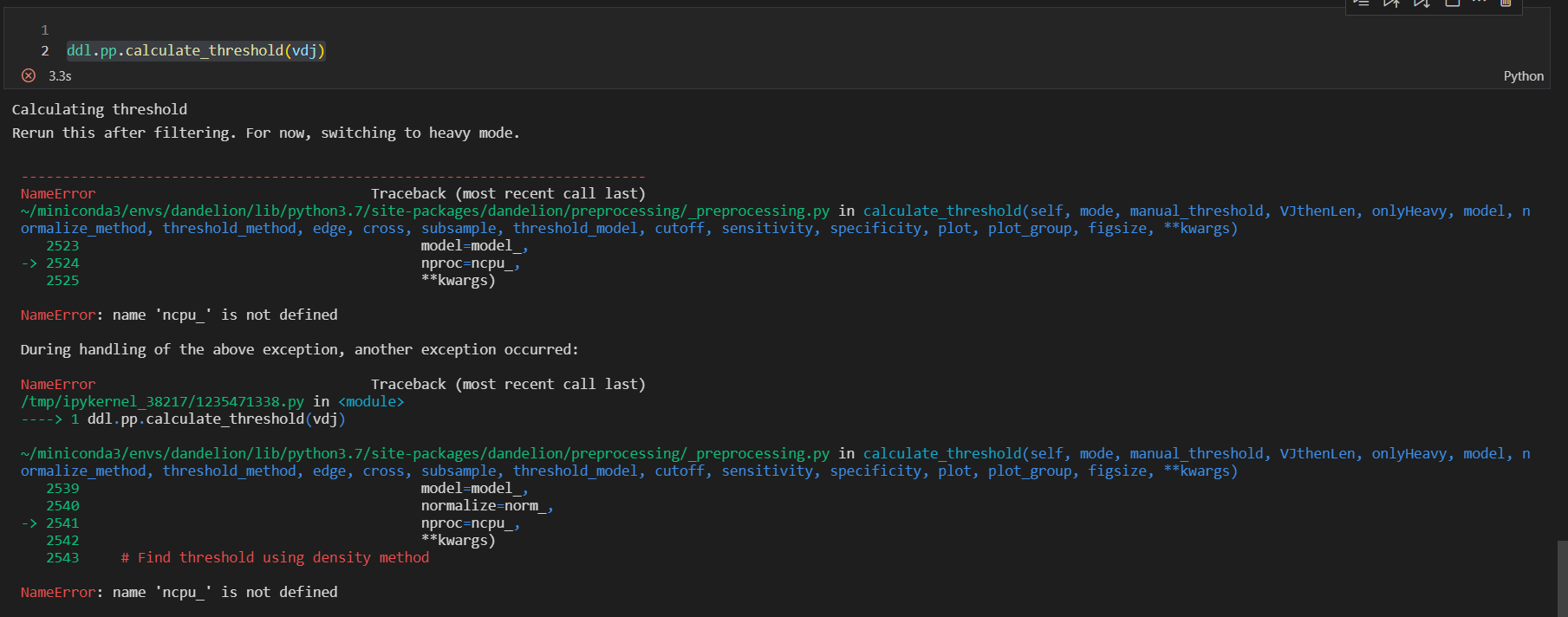
Would you please help me to figure out the problem with BSR Pre_processing. For the step 4 I get this error:
ddl.pp.assign_isotypes(samples, blastdb = "/Users/saramoein/Downloads/dandelion-master/container/database/blast/human/human_BCR_C.fasta")
The error:
FileNotFoundError Traceback (most recent call last)
in
----> 1 ddl.pp.assign_isotypes(samples, blastdb = "/Users/saramoein/Downloads/dandelion-master/container/database/blast/human/human_BCR_C.fasta")
2
3
4
/opt/anaconda3/lib/python3.8/site-packages/dandelion/preprocessing/_preprocessing.py in assign_isotypes(fastas, fileformat, org, correct_c_call, correction_dict, plot, save_plot, show_plot, figsize, blastdb, allele, parallel, ncpu, filename_prefix, verbose)
1004
1005 for i in range(0, len(fastas)):
-> 1006 assign_isotype(fastas[i],
1007 fileformat=fileformat,
1008 org=org,
/opt/anaconda3/lib/python3.8/site-packages/dandelion/preprocessing/_preprocessing.py in assign_isotype(fasta, fileformat, org, correct_c_call, correction_dict, plot, save_plot, show_plot, figsize, blastdb, allele, parallel, ncpu, filename_prefix, verbose)
818 if verbose:
819 print('Running blastn \n')
--> 820 _run_blastn(filePath, blastdb, format_dict[fileformat], org, verbose)
821 # parsing output into a summary.txt file
822 if verbose:
/opt/anaconda3/lib/python3.8/site-packages/dandelion/preprocessing/_preprocessing.py in _run_blastn(fasta, blastdb, fileformat, org, verbose)
416 print('Running command: %s\n' % (' '.join(cmd)))
417 with open(blast_out, 'w') as out:
--> 418 run(cmd, stdout=out, env=env)
419
420 def _parse_BLAST(fasta, fileformat):
/opt/anaconda3/lib/python3.8/subprocess.py in run(input, capture_output, timeout, check, *popenargs, **kwargs)
491 kwargs['stderr'] = PIPE
492
--> 493 with Popen(*popenargs, **kwargs) as process:
494 try:
495 stdout, stderr = process.communicate(input, timeout=timeout)
/opt/anaconda3/lib/python3.8/subprocess.py in init(self, args, bufsize, executable, stdin, stdout, stderr, preexec_fn, close_fds, shell, cwd, env, universal_newlines, startupinfo, creationflags, restore_signals, start_new_session, pass_fds, encoding, errors, text)
856 encoding=encoding, errors=errors)
857
--> 858 self._execute_child(args, executable, preexec_fn, close_fds,
859 pass_fds, cwd, env,
860 startupinfo, creationflags, shell,
/opt/anaconda3/lib/python3.8/subprocess.py in _execute_child(self, args, executable, preexec_fn, close_fds, pass_fds, cwd, env, startupinfo, creationflags, shell, p2cread, p2cwrite, c2pread, c2pwrite, errread, errwrite, restore_signals, start_new_session)
1704 if errno_num != 0:
1705 err_msg = os.strerror(errno_num)
-> 1706 raise child_exception_type(errno_num, err_msg, err_filename)
1707 raise child_exception_type(err_msg)
1708
FileNotFoundError: [Errno 2] No such file or directory: 'blastn'
Thanks