This is the official repository for SlicerMorph project.
Our project aims to enhance the open-source 3D Slicer platform with cutting-edge tools to assist biologists, anthropologists, and morphologists in analyzing 3D data from research imaging modalities. Our ultimate goal is to foster a collaborative community within the 3D Slicer ecosystem to facilitate seamless data exchange and promote the advancement of open science.
SlicerMorph streamlines digital morphology research by enabling effortless data import, visualization, measurement, annotation, and geometric morphometric analysis on 3D data, including volumetric scans (CTs and MRs) and 3D surface scans, all within the 3D Slicer application. Say goodbye to multiple programs, different file formats, and workflows!
SlicerMorph: An open and extensible platform to retrieve, visualize and analyze 3D morphology is now available in Methods in Ecology and Evolution as an open-access paper. A more detailed version of this manuscript that explains all digital anatomy workflows in Slicer is also available as a preprint on Biorxiv.
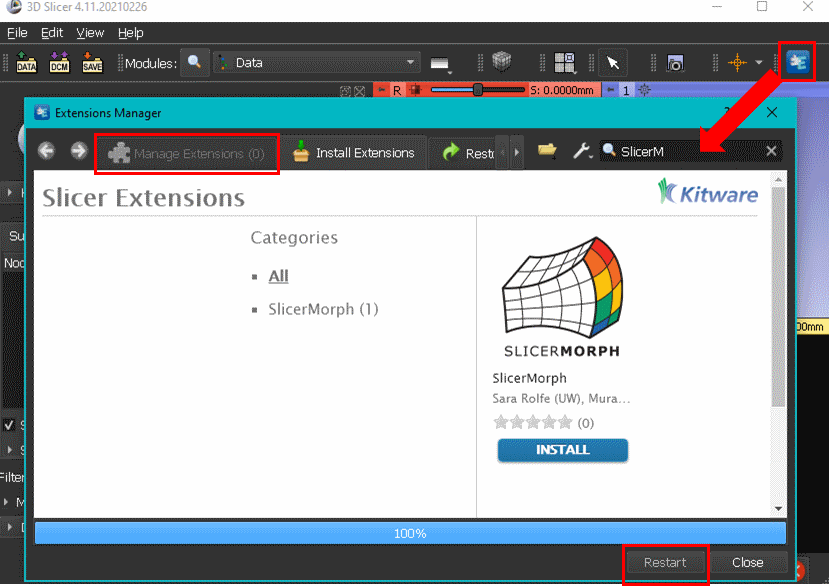
Official method of obtaining SlicerMorph is through extension mechanism of 3D Slicer. To obtain SlicerMorph, please first install the latest stable release (at the moment release 32438, v5.6.1) of 3D Slicer and use the Extensions Manager module to search for and install the SlicerMorph extension. After restarting 3D Slicer, all SlicerMorph modules will be available in the application.
If you use SlicerMorph in your research, please cite one or more of these publications.
-
SlicerMorph as a general platform for digital morphology: Rolfe, S., Pieper, S., Porto, A., Diamond, K., Winchester, J., Shan, S., … Maga, A. M. (2021). SlicerMorph: An open and extensible platform to retrieve, visualize and analyze 3D morphology. Methods in Ecology and Evolution, 12:1816–1825. https://doi.org/10.1111/2041-210X.13669 (Open access)
-
For CreateSemiLMPatches, ProjectSemiLMs, PseudoLMGenerator: Rolfe, S., Davis, C., & Maga, A. M. (2021). Comparing semi-landmarking approaches for analyzing three-dimensional cranial morphology. American Journal of Physical Anthropology. 175(1):227-237. https://doi.org/10.1002/ajpa.24214
-
For ALPACA: Porto, A., Rolfe, S. M., & Maga, A. M. (2021). ALPACA: A fast and accurate approach for automated landmarking of three-dimensional biological structures. Methods in Ecology and Evolution. 12:2129–2144. http://doi.org/10.1111/2041-210X.13689 (Open access).
-
For MALPACA (Multi-Template ALPACA): Zhang, C., Porto, A., Rolfe, S., Kocatulum, A., & Maga, A. M. (2022). Automated landmarking via multiple templates. PLOS ONE. 17(12). e0278035. https://doi.org/10.1371/journal.pone.0278035 (Open access).
To cite 3D Slicer as a general purpose biomedical visualization platform, please use: Kikinis, R., Pieper, S. D., & Vosburgh, K. G. (2014). 3D Slicer: A Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support. In Intraoperative Imaging and Image-Guided Therapy (pp. 277–289). Springer, New York, NY. https://doi.org/10.1007/978-1-4614-7657-3_19
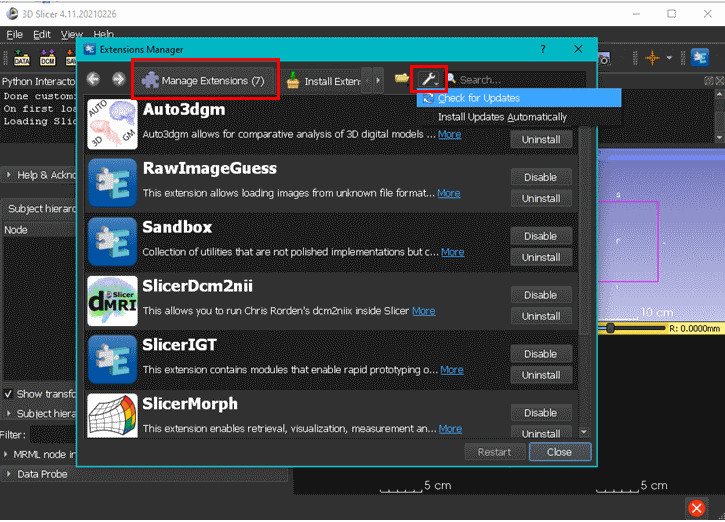
We continuously add new features and functionalities to SlicerMorph packages. To make sure you are using the latest version, go to Extension Manager, click Manage Extensions tab and then use the "Check for updates" function. Once updates are installed, please restart Slicer for changes to take effect. This feature is only available for stable releases. If you are using the preview version, you have to install a newer preview version to obtain the changes introduced to SlicerMorph.
Modules in SlicerMorph are organized in three broad categories:
- ImageStacks: A general purpose tool to import non-DICOM image sequences (png/jpg/bmp/tiff) into Slicer. Provides options to specify voxel size, select image quality (preview, half, full resolution), define an ROI for import, load every Nth slice (skip slice), or reverse stack order (to deal with mirroring of the specimen).
- SkyscanReconImport: Imports an image stack from Bruker/Skyscan reconstruction software (Nrecon) with correct voxel spacing and orientation as a 3D volume. User just needs to point out to the _Rec.log file that is outputted by the Nrecon software. Also possible to process multiple scans through the batch conversion interface (requires all reconstructions to be under a single directory tree)
- GEVolImport: Utility to import the .VOL files generated by the GE/tome microCT scanners by parsing the associated reconstruction file (.PCR).
- MorphoSourceImport: A utility to query and download open-access data from MorphoSource. Requires an API key from MorphoSource.
- GPA: Performs Generalized Procrustes Analysis (GPA) with conducts principle component analysis (PCA) of GPA aligned shape coordinates, provides graphical output of GPA results and real-time 3D visualization of PC warps either using the landmarks of mean shape, or using a reference model that is transformed into the mean shape. Visualization of 3D shape deformation of the reference model can be exported as video clips. The input into the module is a folder path containing a number of landmark files stored in Slicer’s fcsv format and optionally a 3D model and accompanying set of landmarks to be used as reference model in 3D visualization of PCA results.
- CreateSemiLMPatches: Provides triangular patches of semi-landmarks that are constrained by three fixed anatomical landmarks. The input into the module is a 3D model and its accompanying set of fixed landmarks, and users generate and visualize the patches by specifying triplets of fixed landmarks that form a triangle.
- PseudoLMGenerator: This module uses the 3D model’s geometry to create a dense template of pseudo-landmarks. The landmark placement is constrained to the external surface of the mesh. Points sampled on the template are projected to the mesh surface along the surface normals of the template and then filtered to remove those within the sample spacing distance, improving regularity of sampling. This module can be used to generate a large number of points on a 3D model that can serve as a landmark template for additional samples in the dataset.
- ALPACA: Automated Landmarking through Pointcloud Alignment and Correspondence Analysis. ALPACA provides fast landmark transfer from a 3D model and its associated landmark set to target 3D model(s) through pointcloud alignment and deformable model registration. Unlike the two semi-landmarking methods above, it does not require presence of fixed landmarks. Optimal set of parameters that gives the best correspondence can be investigated (and outcome can be visualized) in single alignment mode, and then applied to a number of 3D models in batch mode. Invoked first time, ALPACA needs your permission to download necessary libraries. Depending on the internet speed, download may take sometime but it is a one-time event.
- MarkupEditor: A plugin that enables to select and edit subsets of dense semi-landmarks by drawing an arbitrary closedcurve in the 3D viewer using right-click context menus in 3D viewer. Selected landmarks can be removed from the current node or copied into a new fiducial node. Useful to quickly identify and remove outliers from spherical sampling procedure, or group landmarks into anatomical regions for downstream analyses.
- ImportFromURL: A simple utility that will download the data from the provided URL into the current Slicer scene.
- Animator: A basic keyframe-based animation of 3D volumes. Supports interpolation of regions of interests, rotations, and transfer functions for volume rendering. Output can be either as an image sequence of frames or can be compiled into mp4 format.
- PlaceSemiLMPatches: A utility to apply the generated connectivity table from the CreateSemiLMPatches module to other 3D models with the same set of anatomical landmarks. A new set of semiLandmarks are sampled directly from the new models using the specified connectivity map. Inputs: The connectivity table from CreateSemiLMPatches, a directory with the list of 3D models that semi-landmarks will be placed on, and associated anatomical landmark files (in fcsv format). Both model and its corresponding fcsv file should have the same filename prefix.
- ProjectSemiLM: A utility to transfer a template of semilandmarks to new 3D models using Thin Plate Splines (TPS) warp. Requires existence a set of corresponding anatomical landmarks in the template and target models. Inputs: a base 3D model with its anatomical landmark and the semi-landmark template sets used as reference; a directory with the list of 3D models that semi-landmarks will be projected on and their associated anatomical landmark files (in fcsv format). Both model and its corresponding fcsv file should have the same filename prefix.
- MergeMarkups:. A utility to tag and consolidate fixed and semiLM files into a single merged file.
- MorphologikaLMConverter: Imports a Morphologika formatted landmark file that may contain multiple subjects and exports a Slicer .FCSV landmark file for each individual to a directory specified by the user.
- IDAVLMConverter: Imports IDAV landmark editor files with header length specified by the user.
- ImportSurfaceToSegment: Imports a 3D surface model as a segmentation and prompts the user to edit it using the Segment Editor module.
- SegmentEndoCranium: Automatically segments the endocranial space in a 3D volume of a vertebrate skull. A detailed description of the module and how it works can be found in this tutorial under the automated method.
- SlicerMorph Preferences: Users can choose to enable specific customizations that include keyboard shortcuts, and optimized settings for working with large 3D datasets using the Application Settings menu of 3D Slicer. See the documentation link for list of SlicerMorph specific keyboard shortcuts and modified settings.
- FastModelAlign: Allows users to register a 3D model (source) to a target. Support scaling, rigid and affine transforms.
The following extension is installed automatically with SlicerMorph. Although it is not directly relied on by the modules, it is necessary for many workflows supoorted by SlicerMorph:
The following extension is required for using the SegmentEndocranium. Installation is prompted on opening this module.
We recommend that the user manually install the following extensions that provide useful convenience functions:
- SlicerIGT: Provides landmark driven registration (affine and deformable) of volumes and models.
- Sandbox: Provides utilities like Cross-sectional Area from segments, Colorize Volume, CurvePlanarReformat modules and more.
- DCM2NIIX: Provides a user-interface for the DICOM to NIFTI converter DCM2NIIX. Ideal for stripping metadata from DICOM datasets.
- RawImageGuess: A module that enables the user to import proprietary imaging formats by specifying data type, image dimensions and endianness.
We actively use SlicerMorph to teach a week-long intense short course on 3D Morphometrics and Image analysis. The feedback of the attendees help us prioritize feature development and refinement. We are grateful for the feedback of over 200 participants. Click for contents from the latest workshop, Spring 2022
- SlicerMorph project website: https://slicermorph.github.io (website list upcoming events, including monthly user group meeting. It also has pointers to some of the online specimen repositories).
- SlicerMorph Step-by-Step Tutorials: https://github.com/SlicerMorph/Tutorials
- Please sign up for the SlicerMorph announcement to receive notifications about SlicerMorph project, extension updates and upcoming demos and workshops http://mailman11.u.washington.edu/mailman/listinfo/slicermorph-announcements
- Introduction to Slicer - Official Slicer Documentation
- SlicerMorph YouTube Channel: provides visual tutorials for GPA, semilandmarks, and others.
- Slicer Forum, https://discourse.slicer.org (alternatively you can use your github or google accounts to signup)
This project is supported by NSF Advances in Biological Informatics (1759883) and NSF/DBI Cyberinfrastructure (2301405) grants to A. Murat Maga.
See License.md