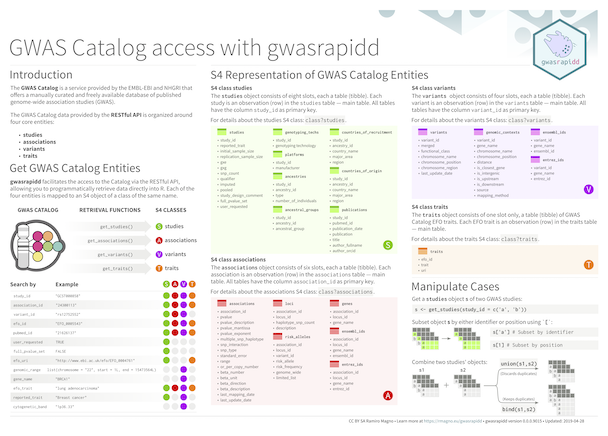
The goal of {gwasrapidd} is to provide programmatic access to the
NHGRI-EBI Catalog of published genome-wide
association studies.
Get started by reading the documentation.
Install {gwasrapidd} from CRAN:
install.packages("gwasrapidd")Get studies related to triple-negative breast cancer:
library(gwasrapidd)
studies <- get_studies(efo_trait = 'triple-negative breast cancer')
studies@studies[1:4]
## # A tibble: 3 × 4
## study_id reported_trait initial_sample_size replication_sample_s…¹
## <chr> <chr> <chr> <chr>
## 1 GCST002305 Breast cancer (estrog… 1,529 European anc… 2,148 European ancest…
## 2 GCST010100 Breast cancer (estrog… 8,602 European anc… <NA>
## 3 GCST90029052 15-year breast cancer… 5,631 European anc… <NA>
## # ℹ abbreviated name: ¹replication_sample_sizeFind associated variants with study GCST002305:
variants <- get_variants(study_id = 'GCST002305')
variants@variants[c('variant_id', 'functional_class')]
## # A tibble: 5 × 2
## variant_id functional_class
## <chr> <chr>
## 1 rs4245739 3_prime_UTR_variant
## 2 rs2363956 missense_variant
## 3 rs10069690 intron_variant
## 4 rs3757318 intron_variant
## 5 rs10771399 intergenic_variant{gwasrapidd} was published in Bioinformatics in 2019:
https://doi.org/10.1093/bioinformatics/btz605.
To generate a citation for this publication from within R:
citation('gwasrapidd')
## To cite gwasrapidd in publications use:
##
## Ramiro Magno, Ana-Teresa Maia, gwasrapidd: an R package to query,
## download and wrangle GWAS Catalog data, Bioinformatics, btz605, 2
## August 2019, Pages 1-2, https://doi.org/10.1093/bioinformatics/btz605
##
## A BibTeX entry for LaTeX users is
##
## @Article{,
## title = {gwasrapidd: an R package to query, download and wrangle GWAS Catalog data},
## author = {Ramiro Magno and Ana-Teresa Maia},
## journal = {Bioinformatics},
## year = {2019},
## pages = {1--2},
## url = {https://doi.org/10.1093/bioinformatics/btz605},
## }Please note that the {gwasrapidd} project is released with a
Contributor Code of
Conduct. By
contributing to this project, you agree to abide by its terms.
- Bioconductor R package gwascat by Vincent J Carey: https://www.bioconductor.org/packages/release/bioc/html/gwascat.html
- Web application PhenoScanner V2 by Mihir A. Kamat, James R. Staley, and others: http://www.phenoscanner.medschl.cam.ac.uk/
- Web application GWEHS: Genome-Wide Effect sizes and Heritability Screener by Eugenio López-Cortegano and Armando Caballero: http://gwehs.uvigo.es/
This work would have not been possible without the precious help from the GWAS Catalog team, particularly Daniel Suveges.