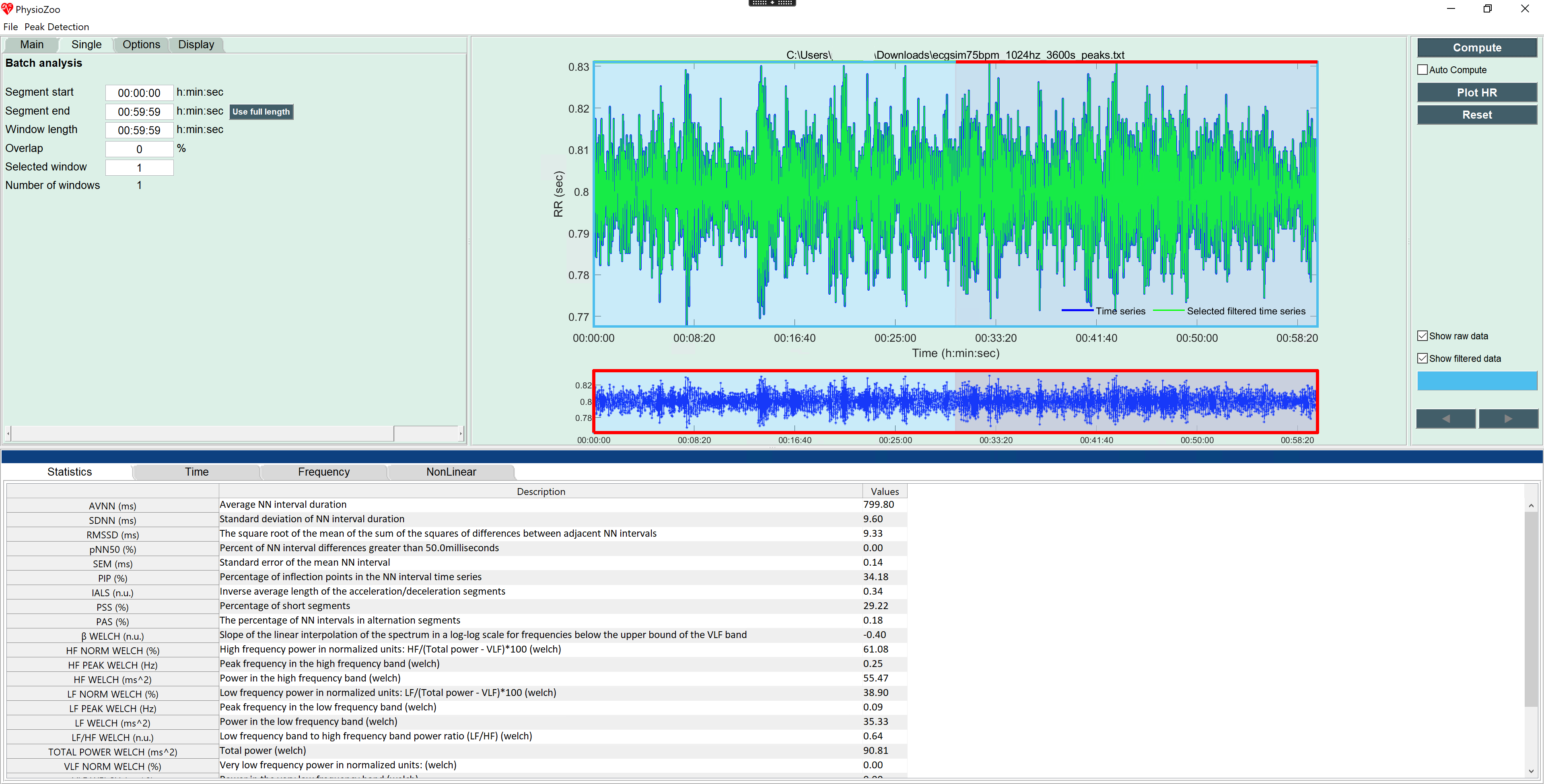
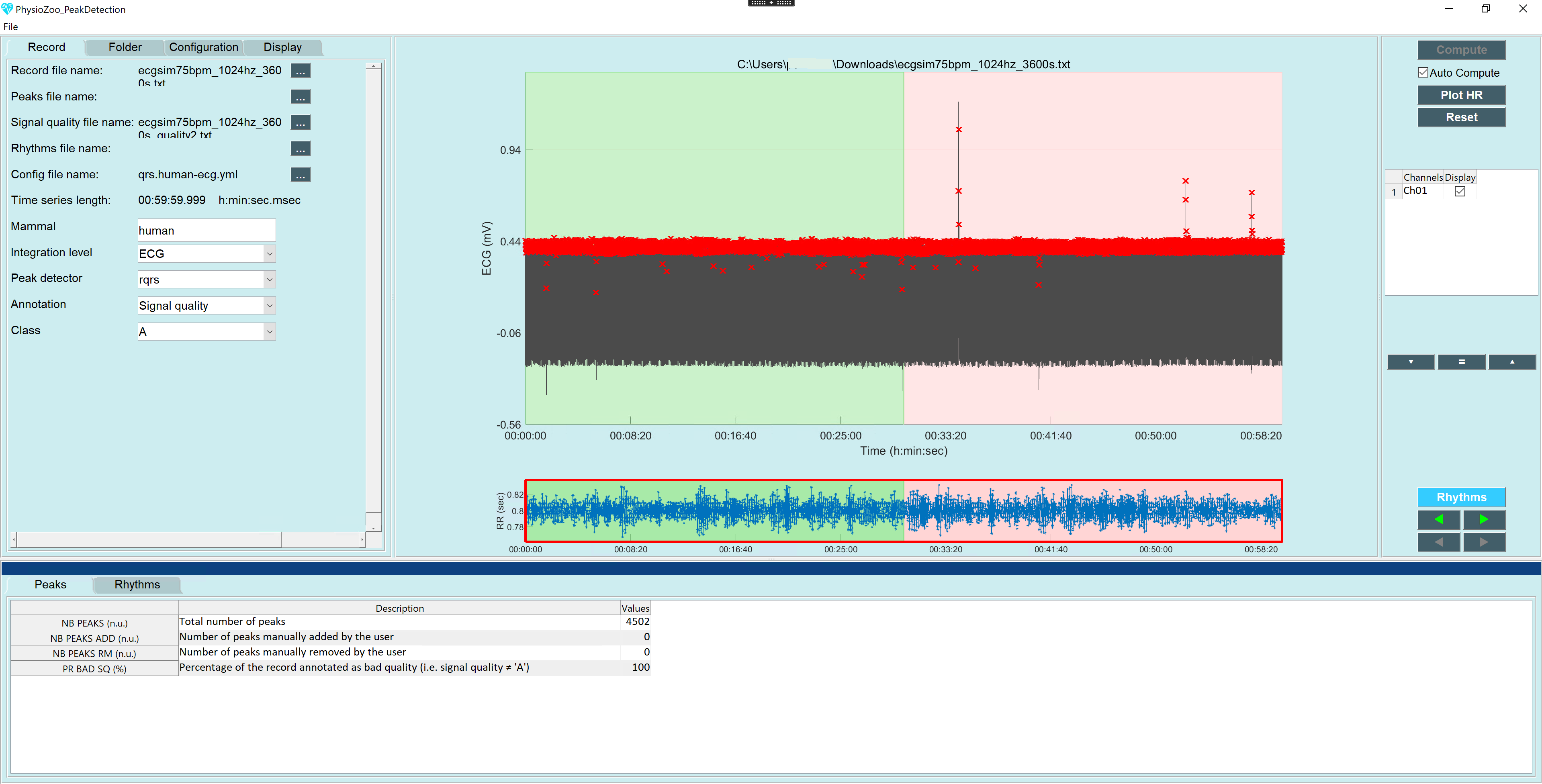
PhysioZoo is a collaborative platform dedicated to the study of the heart rate variability (HRV) from Humans and other mammals’ electrophysiological recordings. The main components of the platform are:
- Software
- An open-source algorithmic toolbox for matlab (
mhrv), which implements all standard HRV analysis algorithms, a selection of peak detection algorithms and prefiltering routines. This can be used within your own data analysis code using themhrvAPI. - An open-source graphical user interface (
PZ-UI) that provides a user friendly interface for advanced HRV analysis of RR-intervals time series and data visualization tools. This enables easy access to HRV analysis without writing any code.
- An open-source algorithmic toolbox for matlab (
- Databases
- A set of annotated databases (
PZ-DB) of electrophysiological signals from different mammals (dog, rabbit and mouse). Available here. - Manually audited peak locations and signal quality annotations for each of the recordings.
- A set of annotated databases (
- Configuration
- A set of configuration files that adapt the HRV measures and
mhrvalgorithms to work with data from different mammals. - All HRV measures can be further adapted for the analysis of other mammals by creating simple human-readable mammal-specific configuration files.
- A set of configuration files that adapt the HRV measures and
The PhysioZoo mission is to standardize and enable the reproducibility of HRV analysis in mammals’ electrophysiological data. This is achieved through its open source code, freely available user interface and open access databases. It also aims to encourage the scientific community to contribute their electrophysiological databases and novel HRV algorithms/analysis tools for advancing the research in the field.
Feedback on how to improve the PhysioZoo platform is welcomed. Do not hesitate to drop us an email at:
Source code, data or interface enhancement contributions are welcome. Look here on how to contribute to PhysioZoo.
Please include the standard citation to PhysioZoo when using the resources available on the platform:
Joachim A. Behar*, Aviv A. Rosenberg*, Ido Weiser-Bitoun, Ori Shemla, Alexandra Alexandrovich, Evgene Konyukhov, Yael Yaniv. 2018. 'PhysioZoo: a novel open access platform for heart rate variability analysis of mammalian electrocardiographic data.' Accepted for publication in Frontiers in Physiology.
*Equal contribution.