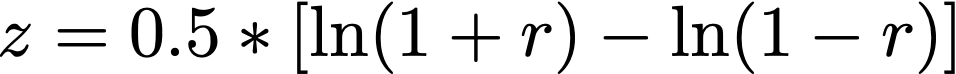merge comparisons (e2r is basically the same as e2c, m2r is the same as m2c(e), the _x2c functions rely on one input whereas the _x2r functions rely on two inputs)
@aaronspring
Regarding comparisons. The six in the code are very similar:
e2r is methodologically the same as e2c
m2r is methodologically the same as m2c and m2e
m2m is a bit outstanding for PMs
The _x2c functions rely on one input whereas the _x2r functions rely on two inputs.
The ref for PMs can easiliy be split from my original ds input.
I would like to bring those concepts together, because then we can combine the compute function. However, then for PPP and NEV PMs would still need an additional argument compute(dp, ref, control) to give the long control run to the function to assess control.std('time') from a long run.
For m2m I would need a serial approach: First to it m2c or m2r and iterate over all members.
@bradyrx
e2r is methodologically the same as e2c
Is there a difference with the supervector approach though? For my e2r I just take the mean of the DPLE if they provide one with a lot of members. Then compare the DPLE one-to-one with the reference, since we expect/force the reference to be the same length as the DPLE.
With your case, the control is much longer than the DPLE, so you have to take a supervector by repeating the DPLE until it is the length of the control, I think?
Same thoughts for the m2r / m2c comparison.
I do think if we combine these into one function, we should use the x2r terminology, since "reference" can capture a control run, assimilation, hindcast, reconstruction, observations, etc.
I would like to bring those concepts together, because then we can combine the compute function.
I think keeping compute_perfect_model and compute_reference separate is probably good so that the user is very explicit on what type of DP they are using. They might accidentally do a PM instead of an assimilation comparison if it's controlled by some single keyword they (or I) forget to turn on or off.
Regardless, if we combine the comparisons into e2r, m2r, etc. we could have a flag for "PM" vs. "reconstruction" that either takes the supervector approach or not. These flags would automatically be run whether compute_perfect_model or compute_reference is run.