navis-org / skeletor Goto Github PK
View Code? Open in Web Editor NEWExtraction of 3D skeletons from meshes.
Home Page: https://navis-org.github.io/skeletor/
License: GNU General Public License v3.0
Extraction of 3D skeletons from meshes.
Home Page: https://navis-org.github.io/skeletor/
License: GNU General Public License v3.0
Is there a reason why:
> mesh = navis.MeshNeuron("meshes/720575940577508022.ply")
> type(mesh1)
Out[5]: navis.core.neurons.MeshNeuron
> fixed = sk.pre.fix_mesh(mesh, remove_disconnected=5, inplace=False)throws silent assertion error?
---> 68 assert isinstance(mesh, tm.Trimesh)
69
70 if not inplace:
AssertionError:
For convenience of using preprocessing functions it would be nice to support MeshNeuron objects.
First, thanks for sharing your code, it's great.
Now my issue: are SWC points supposed to be ordered? Using my data they definitely aren't, and node_ids do not start at 1. Besides that, results look great so I am wondering what I am doing wrong.
This warning: Total face area increased from last iteration. Contraction stopped prematurely after 30 iterations at epsilon 9.6e-06. is displayed during the contraction phase but the contracted mesh looks OK anyway. Any idea?
Note to self: check out https://github.com/nmwsharp/robust-laplacians-py for the mesh contraction.
Hi~ I would like to save the skeleton without the mesh, however I don't know how to do it. Would you please give me some advice? Following are the code I use. Thank you for your kind help.
import open3d as o3d
import matplotlib.pyplot as plt
import numpy as np
import skeletor as sk
import trimesh
import pandas as pd
# reading a mesh to trimesh
mesh_name = '400.stl'
mesh = trimesh.load(mesh_name)
mesh_o3d = o3d.io.read_triangle_mesh(mesh_name)
fixed = sk.pre.fix_mesh(mesh, remove_disconnected=5, inplace=False)
# Contract the mesh
cont = sk.pre.contract(fixed, iter_lim=15)
# Extract the skeleton from the contracted mesh
skel = sk.skeletonize.by_vertex_clusters(cont, sampling_dist=1)
# # Clean up the skeleton
skel_clean = sk.post.clean_up(skel, mesh)
# # Add/update radii
# # swc['radius'] = sk.post.radii(swc, mesh, method='knn', n=5, aggregate='mean')
sk.post.radii(skel_clean, method='knn')
skel.show(mesh=False)
skel.swc.head()
skel.save_swc('save_swc')
print(skel.swc.head())Hi! Thanks for your nice work!
I have some question when testing your algorithm on chair meshes from ShapeNet
I experiment with vertex cluster and edge collapse but neither of them works(what I expect is to extract four legs of the chair).
#!/usr/bin/env python
# coding=utf-8
import os, os.path as osp, shutil
import networkx as nx
import trimesh
import skeletor as sk
import glob
import argparse
parser = argparse.ArgumentParser()
parser.add_argument('--method', type=int, default=0)
args = parser.parse_args()
mesh_files = glob.glob("./samples/*.obj")
for mesh_file in mesh_files:
try:
mesh = trimesh.load(mesh_file)
print('mesh loaded from', mesh_file)
# mesh.show(mesh=True)
# cc = nx.connected_components(mesh.vertex_adjacency_graph)
# print(len(list(cc)), "connected components")
fixed = sk.pre.fix_mesh(mesh, remove_disconnected=0, inplace=False)
cc = nx.connected_components(fixed.vertex_adjacency_graph)
print(len(list(cc)), "connected components after fixing")
# fixed.show(mesh=True)
# fast, work well for tubular meshes(e.g. neurons)
if args.method == 0:
print('wavefront')
skel = sk.skeletonize.by_wavefront(fixed, waves=1, step_size=1)
# fast but needs mesh to be contracted
elif args.method == 1:
print('contraction and by vertex cluster')
fixed = sk.pre.contract()
skel = sk.skeletonize.by_vertex_clusters(fixed, waves=1, step_size=1)
else:
skel = sk.skeletonize.by_edge_collapse(fixed)
skel.show(mesh=True)
shutil.copy(mesh_file, osp.join("./minisamples", osp.basename(mesh_file)))
except:
pass
Did I miss anything important or misunderstand the algorithm? Or is it caused by the bad mesh quality(e.g. non-manifold meshes from ShapeNet)?
Here are some screenshots and the samples along with the script can be found in the attachment.
Appreciate for your help! Wish you a nice day :)
I noticed that sk.skeletonize.by_vertex_clusters can generate KeyErrors when the sampling_dist at certain values like 0.01. Changing the sampling distance to a value that can be represented better by floating point works fine though.
Example Traceback:
Traceback (most recent call last):
File "skeletor_data/contract_mesh.py", line 12, in <module>
skele = sk.skeletonize.by_vertex_clusters(c_mesh, sampling_dist=0.01)
File "skeletor/skeletonize/vertex_cluster.py", line 193, in by_vertex_clusters
vertex_to_node_map = np.array([new_ids[n] for n in vertex_to_node_map])
File "skeletor/skeletonize/vertex_cluster.py", line 193, in <listcomp>
vertex_to_node_map = np.array([new_ids[n] for n in vertex_to_node_map])
KeyError: 413
Hi, I have a set of vertices and faces that I have given to Trimesh (my mesh is a perfect cylinder (all surface are closed)) .
But when I compute the skeletonization, it seems that something wrong has happened. The mesh isn't complete and so the skeletonization didn't work...
What did I do wrong ?
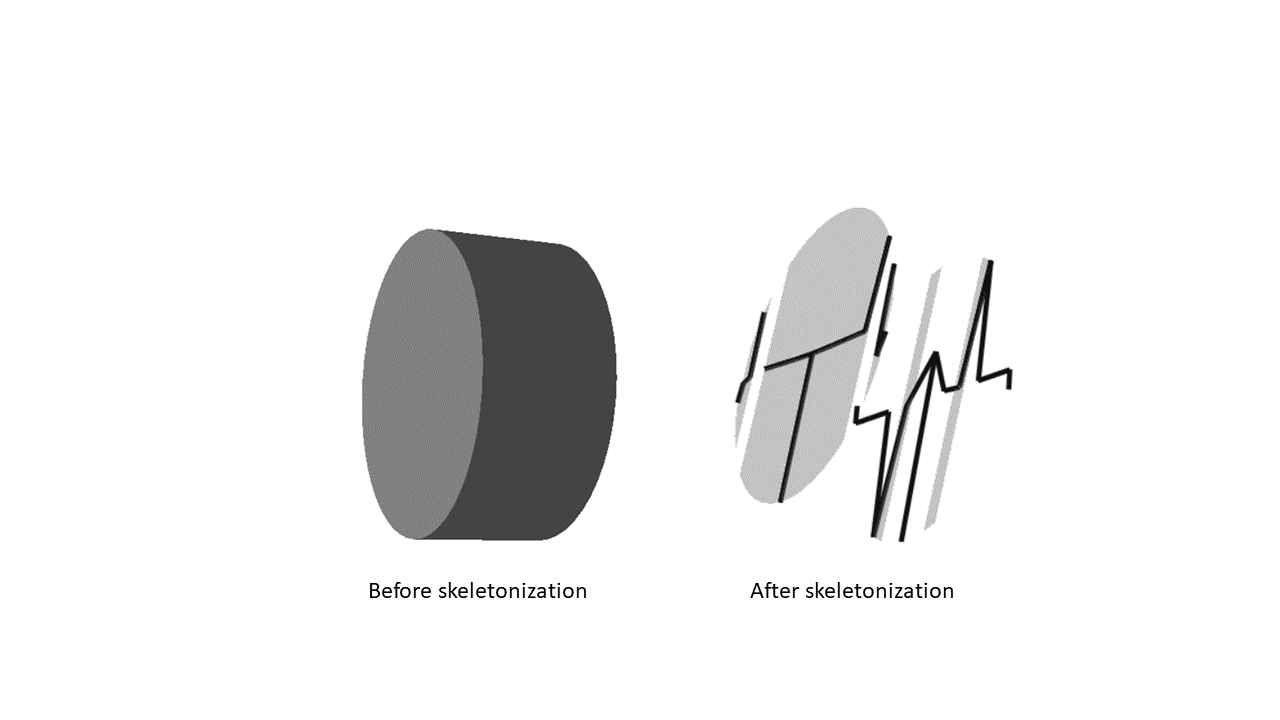
Here is my code for that part :
mesh = trimesh.Trimesh(vertices=Matrixvertices,faces=triangle,process=False)
trimesh.Scene(mesh).show()
fixed = sk.pre.fix_mesh(mesh, remove_disconnected=5, inplace=False)
skel = sk.skeletonize.by_wavefront(fixed, waves=1, step_size=1)
skel.mesh_map
skel.swc.head()
skel.show(mesh=True)
Hi, I am working on the skeleton of a linear structure. Since it has a lot of small branches, I want to prune them and select the largest one. Do you have any recommendation or give me some help about how to do it? The vtk file is attached and you should convert to Trimesh object firstly. By the way, is there a way to output skeleton as a vtk or stl or 2D imagefile? Since I tried to use trimesh.Path3D.exportto output a stl file, but when I open the ParaView to load the path, I cannot see it. Do you have any idea about this issue? I am looking forward to your response, thanks!
Hi @schlegelp ,
I am sure you have come across this skeletonization method. The Mean Curvature Skeletons approach will be an excellent addition to this library. The paper (linked below) says that their system is better than the Skeleton Extraction by Mesh Contraction (Au et al.).
The good news is that this work is based heavily on the Au et al. approach, with the mesh contraction precisely the same according to my observation. They seem to have a better system (easier to implement, presumably :D) in place of the mesh connectivity surgery and embedding refinement used in Au et al.
The paper: http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.261.3158&rep=rep1&type=pdf
There is a C++ implementation of this: https://github.com/taiya/starlab-mcfskel
I hope it works out well! I am enjoying the library! I am using it for non-neuron-based applications, due to which I have to figure out the mechanisms of the skeletonization algorithms. Exciting stuff!
Here I am again,
I wanted to try out the tangent ball method for skeletonization but it doesn't seem to work.
I run the example code:
fixed2 = sk.pre.fix_mesh(aorta_mesh, fix_normals=True, remove_disconnected=10) skel2 = sk.skeletonize.by_tangent_ball(fixed2)
When I run this, I don't get any output and the code doesnt stop running. No progress bar is shown.
Are you familiar with this issue? I think the mesh itself might be the issue but I don't know how to find out.
I've tried other methods using the same mesh and it does work for those meshes however.
Do you know what might be causing this?
Hello, first of all, thank you very much for your efforts and results, so that I successfully achieved the extraction goal of the three-dimensional point cloud tree skeleton, your algorithm is simple and efficient, personally I think it is very good.
But I also encountered a small problem in the process of use, that is, the branches in the model of the original point cloud ply file I entered have some breaks, so I don't want to skeletonize these fractured and incomplete branch parts, of course, I also see that the mesh shrinkage function defines the function of removing the broken part, but after practice, I don't seem to see the application of this parameter in the mesh shrinkage function definition, maybe I made a mistake, and I hope you can give me some advice, Let me successfully achieve only the skeleton process of the complete branch, below is my code and the point cloud after skeletonization, thank you in advance!
Skeletor is relying on Trimesh 's "remove_degenerate_faces()" to deal with degenerate faces, which will only remove faces composed of two vertices, as I understand it. Our neuron meshes are generated with a rather unfortunately large number of t-vertices (3 vertices in a line with a zero area face associated with them). Possible solutions would be to switch to Blender's degenerate dissolve to deal with them, which I believe works pretty well. Meshlab's function for removing them via edge collapse also seems to work for me and is what I have been using. An additional solution may be to roll our own by finding the ones with zero or negligible area (trimesh.base.area_faces or maybe trimesh.base.face_angles ) and then dealing with them by collapsing the edge associated with the middle point? Or by deleting the middle point and then using trimesh.repair.fill_holes(mesh) to put the face back? This would save having to do a type conversion to blender or meshlab and back...
Hi, I am working on the skeleton of 3D mesh. It is good to plot the skeleton path along with 3D mesh. However, if I want to only show the skeleton path, it displays nothing. Here is the screenshot of code and results. Hope someone can help me solve this issue. Also, I attached the mesh vtk file for you to reproduce.
Hello,
I have a non-watertight mesh (OBJ) and the output skeleton is KO (very few points).
Here is my input file:
MeshToSkeletonize.zip
Hello,
I am trying to adapt this library for use with Blender 3d. I am currently working on a project to extract skeletons from a 3D model dataset. However, this dataset I'm using has models with mesh errors that produce ZeroDivision errors when I try to use Py_BL_MeshSkeletonization. Since this is currently the only other public 3D mesh skeletonization library (that I'm aware), I was hoping to use this to extract the remaining skeletons.
I was able to install navis and skeletor inside Blender and the scripts are capable of running. However, my results are less than desired. Is there a way to replicate the TreeNeuron (for the skeleton) in Blender? I understand that navis has a blender3d interface and I've been using the tutorial found here. Also, is there a way to scale the extracted skeleton back to the original trimesh object?
Thank you.
Below is my sample script (insider Blender Python Environment):
import bpy
import trimesh
import skeletor as sk
import os
import numpy as np
import scipy
import ncollpyde
import navis
import navis.interfaces.blender as b3d
# Remove the default cube
bpy.ops.object.select_all(action='DESELECT')
# If default cube is present in scene, remove it
if bpy.data.objects.get("Cube") is not None:
bpy.data.objects['Cube'].select_set(True)
bpy.ops.object.delete()
# Path to model resources obj folder on local machine
obj_parent_dir = "/home/rstewar2/Documents/RPE/AnimSkeleton/ModelResource_Dataset_preprocessed_3DV19/model_resource_data"
obj_dir = os.path.join(obj_parent_dir, "obj")
obj_file_path = os.path.join(obj_dir, "8556.obj")
"""
obj_file = open(obj_file_path)
mesh = trimesh.exchange.obj.load_obj(obj_file, maintain_order=True)
obj_file.close()
"""
#t_mesh = trimesh.Trimesh(vertices=mesh['vertices'],faces=mesh['faces'], vertex_normals=mesh['vertex_normals'])
t_mesh = trimesh.load(obj_file_path)
cont = sk.contract(t_mesh, epsilon=1e-07, iter_lim=10, time_lim=None, precision=1e-8,
SL=2, WH0=1, WL0="auto", operator='umbrella', progress=True,
validate=True)
swc = sk.skeletonize(cont, method='edge_collapse', shape_weight=1, sample_weight=.1, output='both', validate=True)
swc_graph = swc[0]
swc_df = swc[1]
#swc_df = sk.clean(swc_df, mesh)
swc_df['radius'] = sk.radii(swc_df, t_mesh, method='ray', aggregate='mean')
skeleton = navis.TreeNeuron(swc_df.copy(), units='8nm', soma=None)
meshneuron = navis.MeshNeuron(t_mesh, units='8nm')
# Initialize Blender handler
h = b3d.Handler()
# Load neurons into scene
h.add(skeleton, connections=True, neurites=True, soma=False)
This is a picture of the original trimesh model:
This is a picture with the extracted mesh (scaled to 25 in x,y,z directions) next to the original model:
Hey,
I was unable to work with your usage example of the code. Cloudvolume is not a particularly user-friendly library and I wanted a local solution using a stl file. I used trimesh instead of cloudvolume and loaded a file and worked on it directly. I add a short code I built for a skeletal visualization using trimesh and a stl file.
Thank you so much for implementing this article, it has helped me a lot.
Tzabar
https://github.com/TzabarDolev/Mesh_skeltonization/blob/main/skeletor_test.py
It's not as good at preserving the mesh topology but it's decent and potentially much faster (and the Laplacian contraction can be finicky).
Here's the Armadillo mesh (170k vertices) with 100 waves which took about a minute:
Could try out multiple iterations of wave contraction followed by simple vertex clustering.
Hi @tomka and @schlegelp ,
I have an STL file of straight cylinders which I would like to skeletonize. Since they are tubular, I used the wavefront method to achieve the skeleton. You can download the STL file from this link.
https://drive.google.com/file/d/1qfxWzML-2eowcJXYhpm4bcHOMIUumTax/view?usp=sharing
The code I used is:
import trimesh
import skeletor as sk
mesh = trimesh.load_mesh('straight_cylinders.stl')
mesh.show()
skel = sk.skeletonize.by_wavefront(mesh, step_size=1)
# sk.post.radii(skel, mesh, method='ray', aggregate='min', n_rays=2000, projection='tangents', fallback='knn')
skel.show()
The skeleton I am getting is not proper for 4 out of the 5 cylinders. Since I am new to this approach, I am not sure what I am missing. Looking forward to your help.
Hi, thanks for your work!
I am a new at this , and I am trying to get the skeleton for animation from a mesh. I've run this code and get result like the left one in this picture, which have much more joints than I need. How can I get the right result in this picture?
Can you give some advice on this? thank you very much!
Hi
I am just getting into segmentation and mesh analysis.
I found your work really cool. And exactly what I need.
however, after I installed and tried to you run the examples posted I ran into:
module 'skeletor' has no attribute 'contract'
I am using spyder. and I followed the installation instructions that is recommended.
I am sorry for such a basic issue here. it is simple problem I believe.
This is the version installed when i check conda list
skeletor 0.2.11 pypi_0 pypi
Thank you
Some nodes (typically start and end nodes) can be the only ones "hit" by a wave. Consequently, the are moved to their own centre - i.e. do not move at all. We should investigate if these nodes can be better placed by e.g. moving them a certain distance towards their parents.
Hey,
I'm testing out your algorithms on a vascular structure with an aneurysm. All methods I have tried out right now seem to work quite well on narrow vascular structures, but when the radius becomes bigger it seems to go wrong. See this:

Might this be due to a not correct mesh? I do use fix_mesh and contract. When using a smaller value than 0.1 for epsilon in the contract function, most of the times the contraction stops prematurely.
Complete code:
import numpy as np
import pyvista as pv
import tkinter as tk
from tkinter import filedialog
import os
import skeletor as sk
import trimesh as tm
import open3d as o3d
import networkx as nx
root = tk.Tk()
root.withdraw()
root.title('Pick patient data')
file_path = filedialog.askdirectory()
aorta_mesh = tm.load(f"{file_path}\Segment_1.stl")
aorta_mesh_simplified = aorta_mesh.simplify_quadric_decimation(len(aorta_mesh.faces)/10)
fixed2 = sk.pre.fix_mesh(aorta_mesh_simplified, fix_normals=True, remove_disconnected=5, inplace=True)
contracted_mesh = sk.pre.contract(fixed2,epsilon=0.1)
skel = sk.skeletonize.by_wavefront(contracted_mesh, waves=50, step_size=1)
#contracted_mesh = sk.pre.contract(aorta_mesh,epsilon=0.2)
skel2 = sk.skeletonize.by_teasar(contracted_mesh,inv_dist=10)
skel3 = sk.skeletonize.by_tangent_ball(contracted_mesh)
skel4 = sk.skeletonize.by_vertex_clusters(contracted_mesh,sampling_dist=1,cluster_pos='median')
big_plotter = pv.Plotter(shape=(2,2))
big_plotter.subplot(0,0)
big_plotter.add_points(np.array(skel.vertices),color='black')
big_plotter.add_mesh(aorta_mesh_simplified,opacity=0.5)
big_plotter.add_text('Wavefront',position='upper_right',font_size=10)
big_plotter.subplot(0,1)
big_plotter.add_points(np.array(skel2.vertices),color='black')
big_plotter.add_mesh(aorta_mesh_simplified,opacity=0.5)
big_plotter.add_text('TEASAR',position='upper_right',font_size=10)
big_plotter.subplot(1,0)
big_plotter.add_points(np.array(skel3.vertices),color='black')
big_plotter.add_mesh(aorta_mesh_simplified,opacity=0.5)
big_plotter.add_text('Tangent Ball',position='upper_right',font_size=10)
big_plotter.subplot(1,1)
big_plotter.add_points(np.array(skel4.vertices),color='black')
big_plotter.add_mesh(aorta_mesh_simplified,opacity=0.5)
big_plotter.add_text('Vertex Cluster',position='upper_right',font_size=10)
big_plotter.show()
Hi @schlegelp and @tomka ,
I am trying to figure out a meshing pipeline (mesh density and quality) that yields the best results when the wavefront method is used for skeletonization. For that, I think I need to know the behind-the-curtain scenes of the wavefront method.
Could you please tell me the research paper or any literature you used to implement this function?
Thank you!
Hi, thank you for sharing this its very useful! I'm using it to obtain centerlines from coronary artery segmentations.
The way the arteries are structured, I have 2 roots and other nodes are branching from them. The skeleton obtained is exactly how I want, but when I traverse along the path, some nodes which are neither the start nor the end are identified as root or leaf.
Is there a way to ensure these type of nodes are connected to 2 other nodes?
I understand this is more of a graph question rather than a skeleton question.
I have added a figure for your reference. The nodes mentioned are circled in green.

I have been using your algorithm on my laptop and it's working fine, but now im working on a different PC and I can't get it working. I have used your pip3 install skeletor, but i get the error like the title says. It gives the error straight from this line: import skeletor as sk.
I saw an earlier issue with the same problem, but in that problem it was at least possible to run part of the code. So I also can't run print(dir(sk)) print(sk.__version__)
However, I think it automatically shows the available methods:
['__builtins__', '__cached__', '__doc__', '__file__', '__loader__', '__name__', '__package__', '__spec__', 'sk']. As you can see there are no methods that I can use. I'm using python 3.7.3 and pip 24.0. I've already tried to uninstall and reinstall skeletor.
Hopefully you can help me out. Thanks
I am trying to extract the extremities in a skeleton by looking at every node and figuring out whether they are connected to only one other node. Following a suggestion from @schlegelp , I am looking at the graph representation of the skeleton.
I am testing this approach by using the example_mesh. After skeletonization by wavefront, I do:
G=skel.get_graph().to_undirected() for nodes in CC: if len(nodes)==2: print(nodes)
That returns nothing.
Upon inspecting what is in CC, I find that if I do print(nodes) for all nodes in CC, I get this as an output: "{'node_id', 'radius', 'y', 'x', 'parent_id', 'z'}". I was expecting the actual node ids and positions rather than just the strings but I am not an expert in networkx. Is there something wrong or is it completely normal?
The example given in the docs is based on a randomly created mesh which is not helpful for someone who wants to test the skeletonization on his own mesh(es). I think it's good to include an example where a mesh is loaded and skeletonized as this is more of a more generic usage.
To this end, could an example be there to load a mesh so that it could be skeletonized? Because if I use open3d, and load the mesh, it would not get skeletonized. It complains about a package called trimesh.
mesh = make_trimesh(mesh, validate=False)
File "../python3.9/site-packages/skeletor/utilities.py", line 59, in make_trimesh
raise TypeError('Unable to construct a trimesh.Trimesh from object of '
TypeError: Unable to construct a trimesh.Trimesh from object of type "<class 'open3d.cuda.pybind.geometry.TriangleMesh'>"
Apparently, a package called trimesh is being used and that contradicts with open3d meshes. Long story short: How could we even test this on our own mesh?
Hi @schlegelp !
In the documentation for skeletonize.by_edge_collapse, you mentioned that you never got this to work well.
I was wondering if you are working on it to make it faster and more robust?
Thanks!
Hi,
I'm having some issues with plotting the skeleton.
I'm using python 3.9.7 and I've managed to get the skeleton from one of my own .STL files.
`mesh = tm.load_mesh(file_path)
plotter = pv.Plotter()
plotter.add_mesh(mesh)
plotter.show()
#fixed = sk.pre.fix_mesh(mesh, remove_disconnected=5, inplace=False)
skel = sk.skeletonize.by_wavefront(mesh, waves=1, step_size=1)
skel.show(mesh=True)`
This is the original mesh:

This is the result of the skeletonization:

I'm currently having the issues that under a lot of camera angles, the skeleton and mesh are not showing at all. I have to randomly turn the camera and hope they appear. The mesh is also not showing for 99%. I do get this warning from python: UserWarning: Could not set COM MTA mode. Unexpected behavior may occur. warnings.warn("Could not set COM MTA mode. Unexpected behavior may occur.") Is this related to my problem and is there a way to solve it?
Hello,
I searched for 2 full days for python modules which support creating a medial axis of a 3 dimensional mesh. Your repository was the only one I found and it looks quite promising. Unfortunately it doesn't seem to work for me as I expect it - although I tried changing the input parameters:
Also I receive the following error when trying to use the method "edge_collapse" instead of "vertex_clusters": "AttributeError: module 'numpy' has no attribute 'float128'".
Here is the code I used.. What am I doing wrong?
import open3d as o3d
import matplotlib.pyplot as plt
import numpy as np
import skeletor as sk
import trimesh
mesh_name = 'D:/.../Multidirectional_Advanced.stl'
mesh = trimesh.load(mesh_name)
cont = sk.contract(mesh, iter_lim=1000)
swc = sk.skeletonize(cont, method='vertex_clusters', sampling_dist=0.1, output='swc')
swc = sk.clean(swc, mesh)
swc.head()
max_val = max(max(swc.x), max(swc.y), max(swc.z))
fig = plt.figure()
ax = fig.gca(projection='3d')
plt.axis('off')
plt.xlim([-max_val, max_val])
plt.ylim([-max_val, max_val])
ax.set_zlim(-max_val, max_val)
ax.scatter(swc.x, swc.y, swc.z, s=10)
plt.show()
Thanks in advance!
Hi,
I'm currently creating some skeleton objects from meshes. Once I crate an object and print its swc attiribute I get this ->
>>> mesh = tm.load_mesh(inputMeshFile)
>>> skeletonObject = sk.skeletonize.by_teasar(mesh, 0.27)
>>> print(skeletonObject.swc)
node_id parent_id x y z radius
0 0 -1 -1.228041 -0.106300 0.001783 None
1 1 0 -1.223447 -0.108507 -0.003192 None
2 2 4 -1.203667 -0.120944 -0.014145 None
3 3 5 -0.332745 0.222496 1.590621 None
4 4 1 -1.217804 -0.115034 -0.011167 None
.. ... ... ... ... ... ...
655 655 534 0.179983 -1.285225 0.181451 None
656 656 480 -0.408439 -0.766452 0.132349 None
657 657 545 -0.085553 -0.860636 0.359673 None
658 658 659 -0.079773 -0.854532 0.385282 None
659 659 657 -0.081701 -0.857868 0.373008 None
[660 rows x 6 columns]
Everything perfect at this, but I really need something specific to continue my study. I want to make first node (root) in the output as the node with minimum "y" coordinate.
To do that,
I have tried to enter its vertex id to by_teasar method as an argumant and did'nt worked.
Than I have tried to, return the DiGraph created in the by_teasar method, and manuplate it to change root, but couldn't do that too.
>>> mesh = tm.load_mesh(inputMeshFile)
>>> vertex_id_with_min_y = np.argmin(mesh.vertices[:,1])
>>> vertex_id_with_min_y
2063
>>> skeletonObject = sk.skeletonize.by_teasar(mesh, 0.27, root=vertex_id_with_min_y)
>>> print(skeletonObject.swc)
node_id parent_id x y z radius
0 0 -1 -1.230152 -0.106686 0.009829 None
1 1 0 -1.234188 -0.113260 0.011952 None
2 2 3 -0.332745 0.222496 1.590621 None
3 3 68 -0.335910 0.223989 1.586074 None
4 4 1 -1.233198 -0.118613 0.011639 None
.. ... ... ... ... ... ...
624 624 493 0.451694 -1.178653 0.168413 None
625 625 493 0.424917 -1.177674 0.161359 None
626 626 452 -0.030012 -0.926118 0.449399 None
627 627 203 0.364310 -1.045986 -0.306445 None
628 628 584 0.389131 -1.143714 0.084383 None
[629 rows x 6 columns]
So, what can I do to get this swc output with first line includes node with minimum y value.
Hi,
What's the best way to cite the wave fronts method in a paper? I found that all the skeletonization methods in the document have a reference except the wavefront.
Thanks!
Hi, I would like to get skeletons of 3D meshes of neuron segments.
Is it possible to set the distance between nodes to a fixed value (for example 300nm) in skeletonizing? Or any post-process should be done.
Thank you!
Hey,
I wanted to try the skeletonize.by_teasar() method but I have no clue what value to put for the inv_dist parameter.
I'm currently using a value of 1 and get the following result, which doesn't seem right:
It looks exactly like the surface of my mesh. I used the following code:
`mesh = tm.load_mesh(file_path)
fixed = sk.pre.fix_mesh(mesh, remove_disconnected=5, inplace=False)
skel_teasar = sk.skeletonize.by_teasar(fixed,inv_dist=1)
skel_teasar.show()`
Hi @schlegelp !
I am building an application that requires the conversion of the skeleton to a networkx object. For that, I am using the navis module as per #19.
Now when I use pyinstaller, I get the error:
FileNotFoundError: [WinError 3] The system cannot find the path specified: 'C:\\Users\\...\\AppData\\Local\\Temp\\_MEI46802\\navis\\data\\gml'
I am using python 3.7 on Windows 10. I don't know if this is a pyinstaller issue, but any input would be really helpful.
I generated the spec file and added the binaries, hiddenimports and hookspath:
# -*- mode: python ; coding: utf-8 -*-
from PyInstaller.utils.hooks import collect_dynamic_libs
block_cipher = None
a = Analysis(['main_script_name.py'],
pathex=['C:\\Users\\<location>'],
binaries=collect_dynamic_libs("rtree"),
datas=[],
hiddenimports=['navis','vtkmodules',
'vtkmodules.all',
'vtkmodules.qt.QVTKRenderWindowInteractor',
'vtkmodules.util',
'vtkmodules.util.numpy_support',
'vtkmodules.numpy_interface.dataset_adapter',],
hookspath=['hooks'],
hooksconfig={},
runtime_hooks=[],
excludes=[],
win_no_prefer_redirects=False,
win_private_assemblies=False,
cipher=block_cipher,
noarchive=False)
pyz = PYZ(a.pure, a.zipped_data,
cipher=block_cipher)
exe = EXE(pyz,
a.scripts,
a.binaries,
a.zipfiles,
a.datas,
[],
name='main_script_name',
debug=False,
bootloader_ignore_signals=False,
strip=False,
upx=True,
upx_exclude=[],
runtime_tmpdir=None,
console=True,
disable_windowed_traceback=False,
target_arch=None,
codesign_identity=None,
entitlements_file=None )
The hooks file contains:
from PyInstaller.utils.hooks import collect_data_files
datas = collect_data_files('trimesh')
I tried adding an additional line to the hooks file to append navis data file; but it still caused the FileNotFoundError
# datas += collect_data_files('navis')
Could you please help me out here? Thanks a lot!
Hello! Thank you for this library.
Is there a way to extract the radius of the minimum/maximum inscribed spheres along the skeleton path?
Thank you!
Hey, as you have developed this method to extract the skeleton by vertex clustering, I would imagine you have read some research paper about this and then implemented it yourself.
I couldn't find any references for it on the home page, and have found only paper, which is this one:
'Example-Based Skeleton Extraction' by S. Schaefer and C. Yuksel.
Do you still know on basis of what research you have developed this code?
Thanks in advance!
Hi !
I'm currently implementing the exact same solution, but on C++ using LibIGL, and there's a couple of questions I have, if you can help me it would mean a lot
Thanks !
Hi @schlegelp !
I am trying to convert a skeleton to a networkx graph upon which I can then use many graph-based algorithms such as shortest path (by assigning the distance between nodes as edge-weights), etc.
Before, I dig deep into this, I wanted to know if this is already done or is there a straightforward way within skeletor to convert a skeleton to a networkx graph? Any input will be very helpful.
I am really enjoying this library by the way!
Thanks!
Upon using teasar skeletonization on Python 3.9.6, one faces the following error:
In [11]: eids = SG.get_eids(path=path, directed=False)
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In [11], line 1
----> 1 eids = SG.get_eids(path=path, directed=False)
TypeError: 'path' is an invalid keyword argument for this function
Any workaround one may know, apart from upgrading the Python version?
Hello, I downloaded your code on the website, but I don't know how to run it. I still don't understand the principle. Can you explain how to run the code?
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.