This package estimates linear models with high dimensional categorical variables and/or instrumental variables.
The package is registered in the General registry and so can be installed at the REPL with ] add FixedEffectModels.
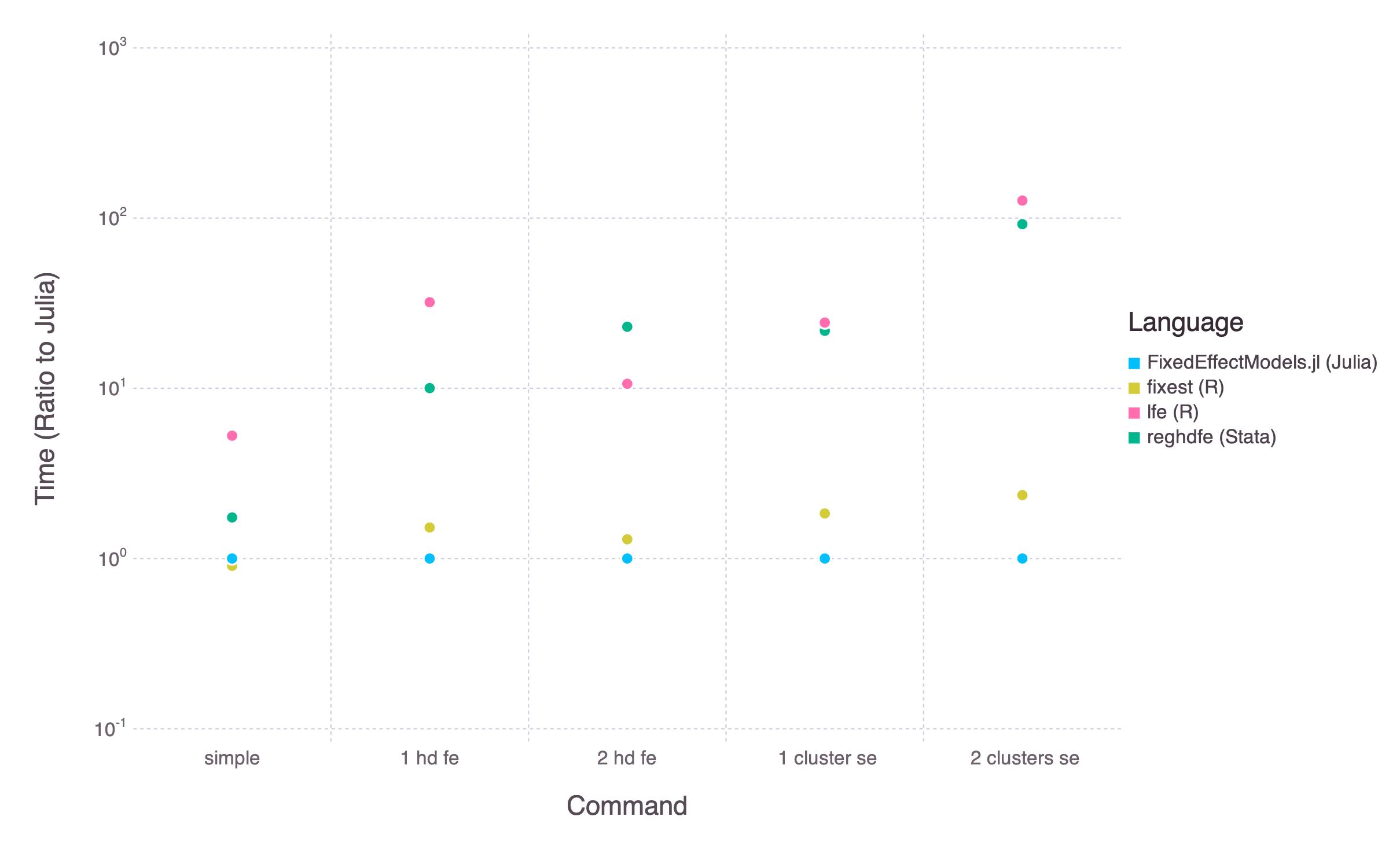
The objective of the package is similar to the Stata command reghdfe and the R packages lfe and fixest. The package is much faster than reghdfe or lfe. It also tends to be a bit faster than the more recent fixest (depending on the exact command). For complicated models, FixedEffectModels can also run on Nvidia GPUs for even faster performances (see below)
using DataFrames, RDatasets, FixedEffectModels
df = dataset("plm", "Cigar")
reg(df, @formula(Sales ~ NDI + fe(State) + fe(Year)), Vcov.cluster(:State), weights = :Pop)
# FixedEffectModel
# =========================================================================
# Number of obs: 1380 Converged: true
# dof (model): 1 dof (residuals): 45
# R²: 0.803 R² adjusted: 0.798
# F-statistic: 13.3382 P-value: 0.001
# R² within: 0.139 Iterations: 5
# =========================================================================
# Estimate Std. Error t-stat Pr(>|t|) Lower 95% Upper 95%
# ─────────────────────────────────────────────────────────────────────────
# NDI -0.00526264 0.00144097 -3.65216 0.0007 -0.0081649 -0.00236038
# =========================================================================-
A typical formula is composed of one dependent variable, exogenous variables, endogenous variables, instrumental variables, and a set of high-dimensional fixed effects.
dependent variable ~ exogenous variables + (endogenous variables ~ instrumental variables) + fe(fixedeffect variable)
High-dimensional fixed effect variables are indicated with the function
fe. You can add an arbitrary number of high dimensional fixed effects, separated with+. You can also interact fixed effects using&or*.For instance, to add state fixed effects use
fe(State). To add both state and year fixed effects, usefe(State) + fe(Year). To add state-year fixed effects, usefe(State)&fe(Year). To add state specific slopes for year, usefe(State)&Year. To add both state fixed-effects and state specific slopes for year usefe(State)*Year.reg(df, @formula(Sales ~ Price + fe(State) + fe(Year))) reg(df, @formula(Sales ~ NDI + fe(State) + fe(State)&Year)) reg(df, @formula(Sales ~ NDI + fe(State)&fe(Year))) # for illustration only (this will not run here) reg(df, @formula(Sales ~ (Price ~ Pimin)))
To construct formula programmatically, use
reg(df, term(:Sales) ~ term(:NDI) + fe(:State) + fe(:Year))
-
The option
contrastsspecifies that a column should be understood as a set of dummy variables:reg(df, @formula(Sales ~ Price + Year); contrasts = Dict(:Year => DummyCoding()))
You can specify different base levels
reg(df, @formula(Sales ~ Price + Year); contrasts = Dict(:Year => DummyCoding(base = 80)))
-
The option
weightsspecifies a variable for weightsweights = :Pop
-
Standard errors are indicated with the prefix
Vcov(with the package Vcov)Vcov.robust() Vcov.cluster(:State) Vcov.cluster(:State, :Year)
-
The option
savecan be set to one of the following::none(default) to save nothing,:residualsto save residuals,:feto save fixed effects, and:allto save both. Once saved, they can then be accessed usingresiduals(m)orfe(m)wheremis the estimated model (the object returned by the functionreg). Both residuals and fixed effects are aligned with the original dataframe used to estimate the model. -
The option
methodcan be set to one of the following::cpu,:CUDA, or:Metal(see Performances below).
reg returns a light object. It is composed of
- the vector of coefficients & the covariance matrix (use
coef,coefnames,vcovon the output ofreg) - a boolean vector reporting rows used in the estimation
- a set of scalars (number of observations, the degree of freedoms, r2, etc)
Methods such as predict, residuals are still defined but require to specify a dataframe as a second argument. The problematic size of lm and glm models in R or Julia is discussed here, here, here here (and for absurd consequences, here and there).
You may use RegressionTables.jl to get publication-quality regression tables.
FixedEffectModels is multi-threaded. Use the option nthreads to select the number of threads to use in the estimation (defaults to Threads.nthreads()).
The package has an experimental support for GPUs. This can make the package an order of magnitude faster for complicated problems.
If you have a Nvidia GPU, run using CUDA before using FixedEffectModels. Then, estimate a model with method = :CUDA.
using CUDA, FixedEffectModels
@assert CUDA.functional()
df = dataset("plm", "Cigar")
reg(df, @formula(Sales ~ NDI + fe(State) + fe(Year)), method = :CUDA)The package also supports Apple GPUs with Metal.jl, although I could not find a way to get better performance
using Metal, FixedEffectModels
@assert Metal.functional()
df = dataset("plm", "Cigar")
reg(df, @formula(Sales ~ NDI + fe(State) + fe(Year)), method = :Metal)Denote the model y = X β + D θ + e where X is a matrix with few columns and D is the design matrix from categorical variables. Estimates for β, along with their standard errors, are obtained in two steps:
y, Xare regressed onDusing the package FixedEffects.jl- Estimates for
β, along with their standard errors, are obtained by regressing the projectedyon the projectedX(an application of the Frisch Waugh-Lovell Theorem) - With the option
save = true, estimates for the high dimensional fixed effects are obtained after regressing the residuals of the full model minus the residuals of the partialed out models onDusing the package FixedEffects.jl
Baum, C. and Schaffer, M. (2013) AVAR: Stata module to perform asymptotic covariance estimation for iid and non-iid data robust to heteroskedasticity, autocorrelation, 1- and 2-way clustering, and common cross-panel autocorrelated disturbances. Statistical Software Components, Boston College Department of Economics.
Correia, S. (2014) REGHDFE: Stata module to perform linear or instrumental-variable regression absorbing any number of high-dimensional fixed effects. Statistical Software Components, Boston College Department of Economics.
Fong, DC. and Saunders, M. (2011) LSMR: An Iterative Algorithm for Sparse Least-Squares Problems. SIAM Journal on Scientific Computing
Gaure, S. (2013) OLS with Multiple High Dimensional Category Variables. Computational Statistics and Data Analysis