A series of MATLAB program which can accelerate the processing of MR images.
Attention! To use those toolkit, an MATLAB toolbox name NIfTI should be set as a path of MATLAB software.£¨NIfTI_20140122 folder is the toolbox for you£©
1.Auto_T1_T2_DWI_selection.m Classify the raw MRI data into T1, T2 and DWI by the information of raw data.
2.autoDWIsprlitbytime.m Useless.
3.DiComDifferenceExam.m Exam the difference between different DiCom files.
4.DTI_bvec_DiffutionToolkit_allfilesread.m Convert all the b vector files into a form which can be used by diffusion toolkit software.
5.rotate_for_trackvis.m Extract special ROI template information for TrackVis software.
6.SeparationByScanningDate.m Separate the Dicom raw data into different groups by the scanning date.
7.standard_space_sorting.m Sorting the output files of PANDA software, which scanning special fields such as 'FA', 'LDHs', 'LDHk'and save them to a mat, then export them into an excel.
8.Mean_FA.m For the calculation of Mean FA value.
9.CIN_network_extraction_toolkit.m Extraction CIN(lable 31:36 in AAL 90 atlas set)to show the construction
10.CIN_network_extraction_toolkit_with_split.m Extract the FA brain networks which are not ioslated from other nodes.
11.img2txt.m, img2txt_new_imgshow, img2txt_new_txtchech.m All of them are used for the construction of VR brain which makes the preparation for the Unity softwares.
12.Network_sorting.m Sorting the output files of Networks of PANDA software.
13.standard_space_sorting.m Sorting the standard space output files of PANDA software.
14.level5_to_level3.m Change the information of ROIs in JHU T1 286 images into level3, which compact them into 52 regions and other named 'xxxxx' regions with 0 value in the lookup table.
15.ppg_peakdetect.m PPGsorting programme of GongPing
16.ImgROIselection.m Select all the ROIs in segmented T1 image and save their location (x,y,z) in txt files.
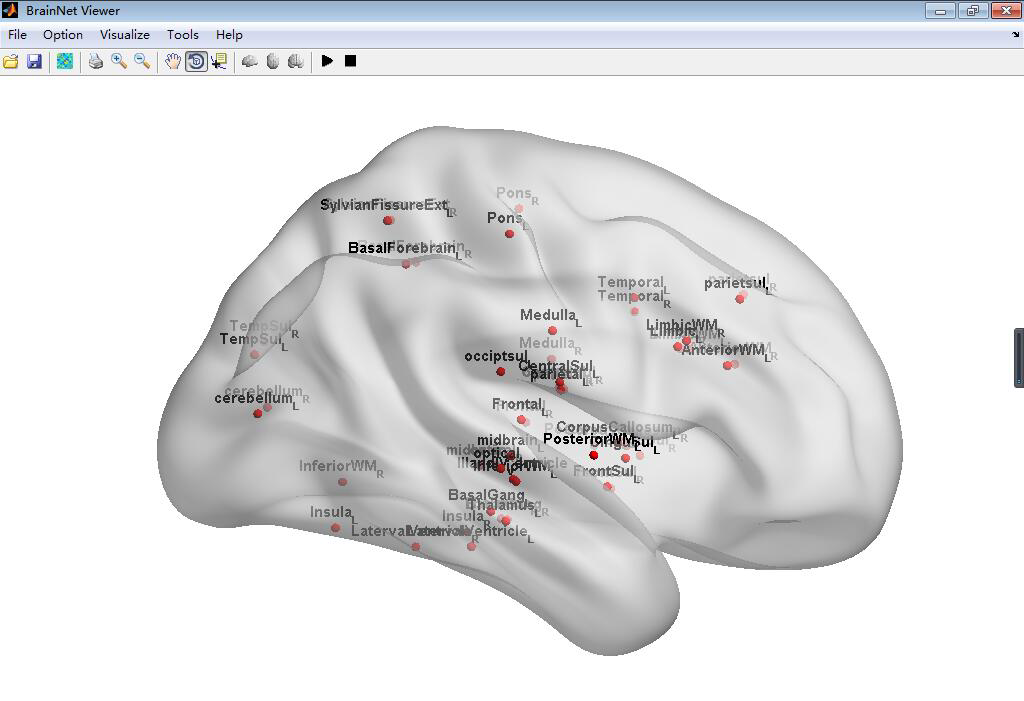
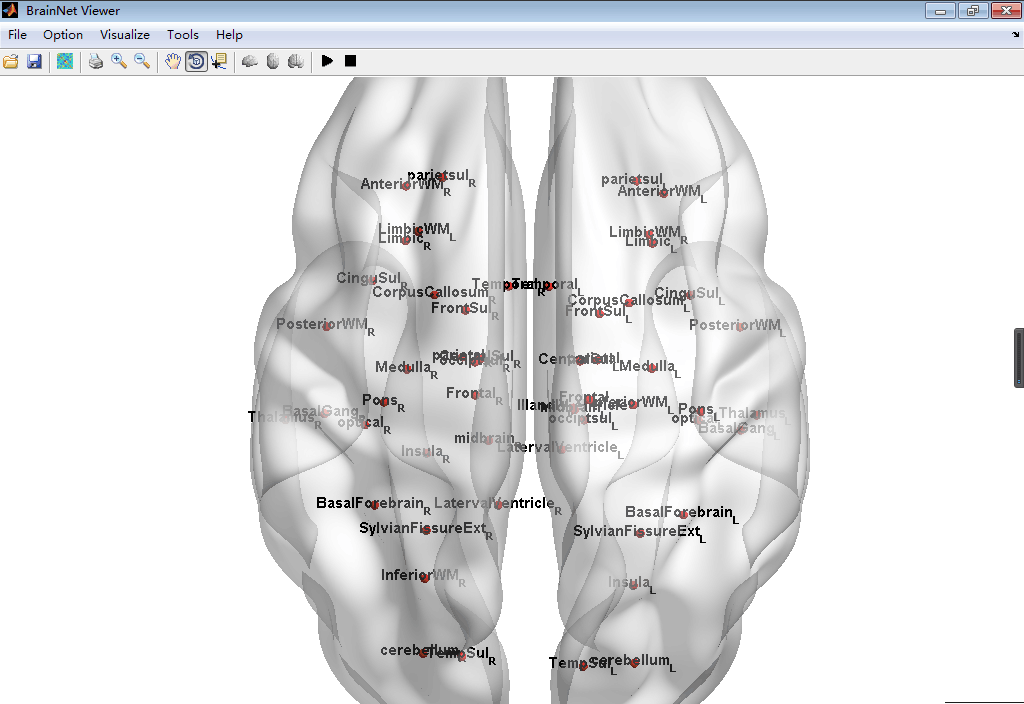
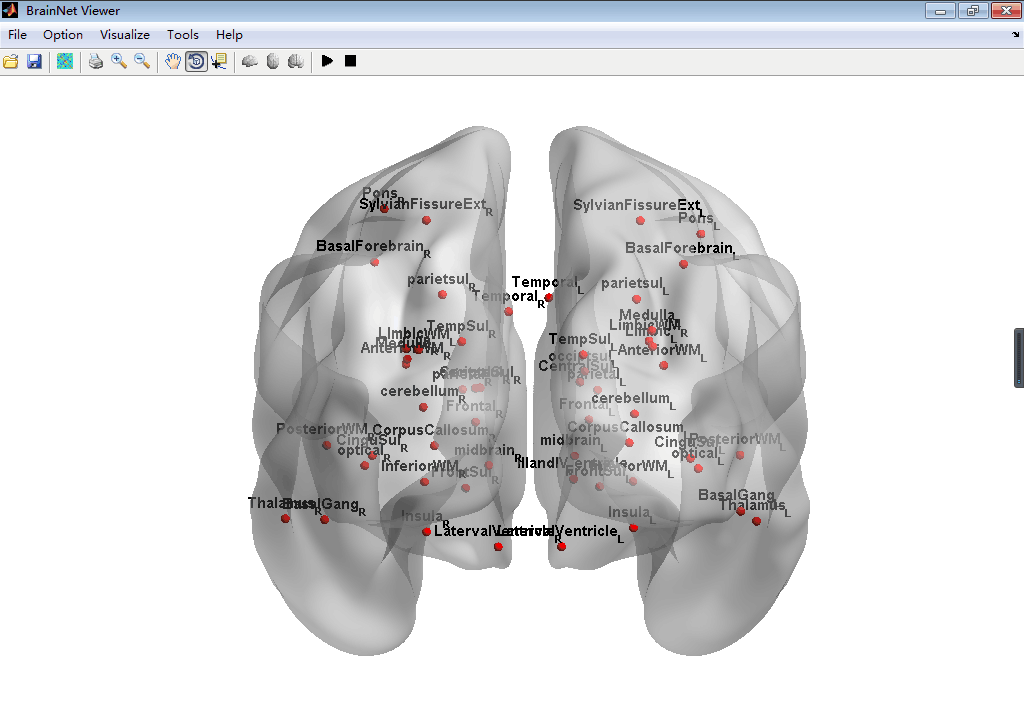
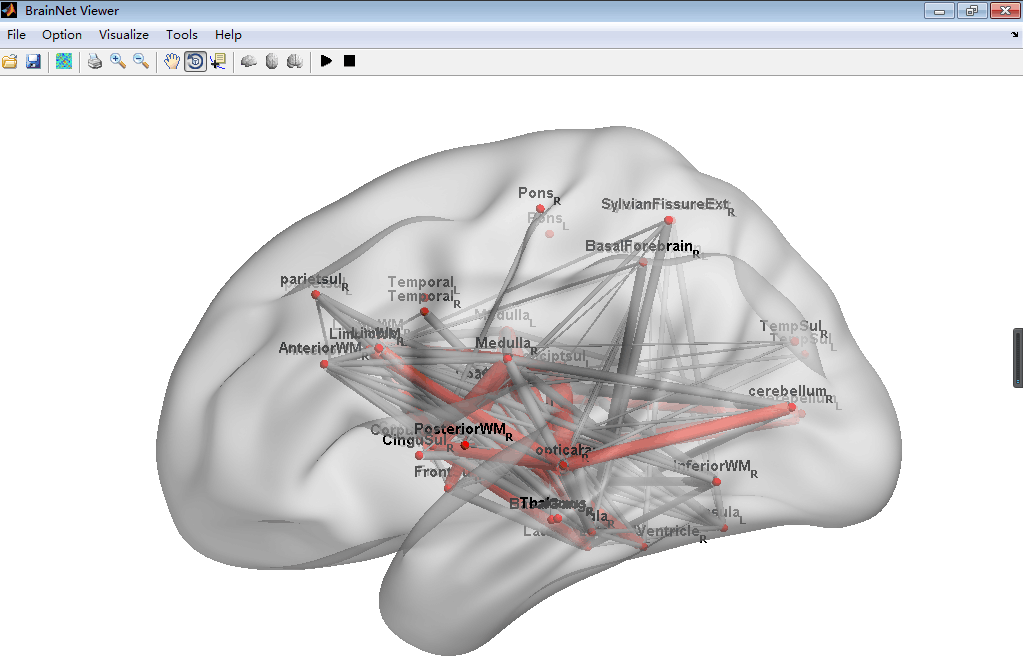
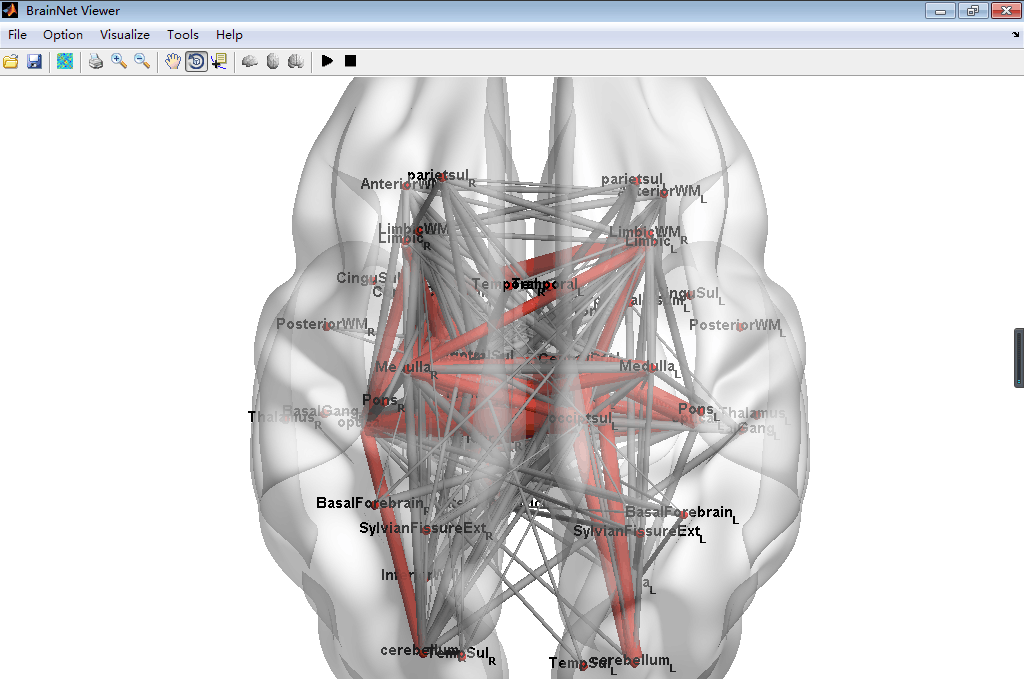
#BrainNetViewer\JHU_level3_53.node
##Install
This node is defined as a new node information can be input in the BrainNetViewer, which is a common used toolbox in MATLAB to visualize the Brain Graph. This node file derived from the brain paracellation result of Johns Hopkins University one lab's result can set in level3 which has 53 brain regions. To install this node file, just place it anywhere you want then load it in the BrainNetViewer.
##Screenshot