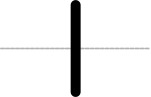
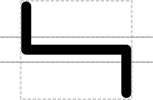
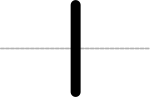
SEP V004: New Glyph Collection
| SEP |
|
| Authors |
Jacob Beal ([email protected]) |
| Editor |
|
| Type |
Specification |
| SBOL Visual Version |
1.1 |
| Status |
Draft |
| Created |
22-Aug-2017 |
| Last modified |
18-Sep-2017 |
Abstract
A number of new glyphs have been proposed over the past few years, and we need to put them to an up-or-down vote.
There are twelve proposals currently pending: Aptamer, Codon, Homology Region, Inverter, Non-Coding RNA, ORI-T, polyA Site, Protein Domain, Specific Recombination Site, Non Directional Sticky End, Tag, Transcript Region
Table of Contents
1. Rationale
Each glyph detailed below in its specification has been provided with an individual rationale for that glyph. Examples are also embedded within each proposal.
2. Specification
Aptamer
Associated SO term(s)
SO:0000031: Aptamer
Recommended Glyph and Alternates
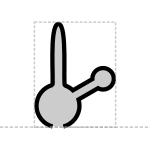
The proposed aptamer glyph is a cartoon diagram of nucleic acid secondary structure like that found in aptamers:
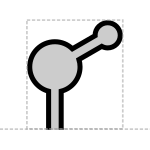
Prototypical Example
theophylline aptamer
Non-Coding RNA Gene
Associated SO term(s)
SO:0001263: Non-Coding RNA Gene
SO:0000834: Mature Transcript Region
Recommended Glyph and Alternates
Two of the proposed non-coding RNA glyphs are both single-stranded RNA "wiggles," one on top of a box:
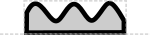
another hovering above the backbone:

One or the other of these should be chosen, but not both.
Prototypical Example
gRNA
ORI-T
Associated SO term(s)
SO:0000724: Origin of Transfer
Recommended Glyph and Alternates
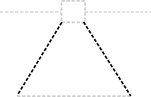
The origin of transfer glyph is circular like Origin of Replication, but also includes an outbound arrow:

Prototypical Example
oriT
Notes
The recommended backbone location of Origin of Replication is not yet fixed; the backbone location of this glyph is intended to match Origin of Replication, so it that is recommended to become below the glyph, this backbone location will shift as well.
polyA site
Associated SO term(s)
SO:0000553: polyA Site
Recommended Glyph and Alternates
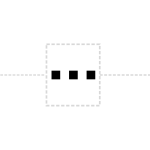
The polyA site glyph is a sequence of As sitting atop the backbone:
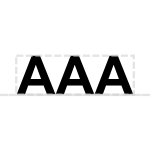
Prototypical Example
polyA tail on mammalian coding sequence
Specific Recombination Site
Associated SO term(s)
SO:0000299: Specific Recombination Site
Recommended Glyph and Alternates
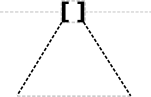
The specific recombination site glyph is a triangle, centered on the backbone, as has appeared in a number of recombinase circuit papers:

Prototypical Example
flippase recognition target (FRT) site
Notes
Potential conflict with proposed Inverter glyph.
3. Examples
See examples in individual glyph proposals.
4. Backwards Compatibility
All proposals are for new glyphs that do not conflict with existing glyphs. Note that two proposals (Inverter and Recombinase Site) do conflict with one another.
5. Discussion
The following proposed options have been considered, but do not have strong support and are thus being removed from consideration unless they pick up significant advocacy. They may be revisited in the future.
Aptamer
Codon
Associated SO term(s)
SO:0000360: Codon
SO:0000318: Start Codon
SO:0000319: Stop Codon
Recommended Glyph and Alternates
The proposed aptamer glyphs are two versions of a cartoon diagram of nucleic acid secondary structure like that found in aptamers:
Nucleotides can be indicated with colors or letters in the boxes:


Proteins can be indicated by a letter above:
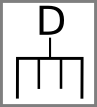
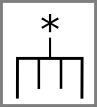
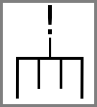
Stop and start codons might be indicated by special symbols:
.png)
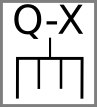
Edits can be indicated by changes:
.png)


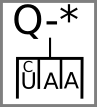

Prototypical Example
UGA stop codon
Notes
If accepted, there will need to be additional work done to elaborate the full specification.
Homology Region
Associated SO term(s)
SO:0000853
Recommended Glyph and Alternates
The homology region glyph is a stretched hexagon hovering above the backbone:

Prototypical Example
Needs a good example
Inverter
Associated SO term(s)
No SO term currently exists
Recommended Glyph and Alternates
The inverter glyph is a triangle, echoing the buffer glyph from electronics. It might be either above or on the backbone.


Prototypical Example
Needs a good example
Notes
Potential conflict with proposed Specific Recombination Site glyph.
Non-Coding RNA
Squiggle with teeth:

Peeling comb suggesting an RNA sequence partially attached to the backbone:

Non Directional Sticky End
Associated SO term(s)
SO:0001692 (unspecified direction)
Recommended Glyph and Alternates
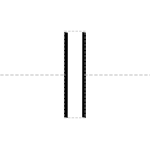
A sticky restriction site of unspecified direction is an angled set of cuts:
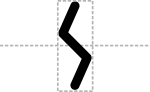
Prototypical Example
EcoRI restriction site.
ORI-T
Spirals outward toward a new destination rather than being a closed circle. Two slightly different variants of spiral are proposed for consideration:
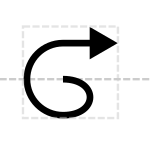

Protein Domain
Associated SO term(s)
SO:0000417 Polypeptide Domain
Recommended Glyph and Alternates
A number of proposals have been made for Protein Domain glyphs. These are:
- A chevron, which composes nicely with the standard CDS pentagon

- A plain rectangle (the same as user defined):

- A chevron with a vertical break:


- A broken arrow (i.e., broken alternate CDS)

- A CDS pentagon with a chevron break:

Prototypical Example
VP64 activation domain
Notes
Protein domain should have the same recommended vertical position as CDS, but CDS does not have a recommended vertical position yet, so these proposals do not either.
Tag
Associated SO term(s)
SO:0000324: Tag
Recommended Glyph and Alternates
The tag glyph is a diagonal rectangle with clipped corners, reminiscent of a stereotypical paper gift tag:
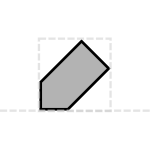
Prototypical Example
PEST tag
Copyright

To the extent possible under law,
SBOL developers
has waived all copyright and related or neighboring rights to
SEP V004.
This work is published from:
United States.










































.png)

.png)