Common Lisp Implementation of NeuroEvolution of Augmenting Topologies (NEAT)
(scroll to the bottom for demonstration runs)
The Lisp package described here was developed according to a technique devised by Stanley and Miikkulainen in a paper called Evolving Neural Networks through Augmenting Topologies. A copy of the paper can be obtained here: http://nn.cs.utexas.edu/downloads/papers/stanley.ec02.pdf
Please note that the purpose of this documentation is not to describe how NEAT works, but rather to:
- Explain how to use the package,
- Point out the deviations from the original paper, and
- Describe the file structure of the package.
Example client code: xor-experiment-parameters.lisp and xor-test.lisp.
- Neural networks do not have loops. The connections in the starter genome should therefore create no loops either.
- Number of neural network inputs and outputs for any given experiment does not change.
- Fitness is positive or zero. I do not remember why this is the case, so maybe the fitness could be negative.
- Load experiment parameters file (e.g. xor-experiment-parameters.lisp).
- Load the package:
(ql:quickload :cl-neat)
Loading through asdf probably works too. 3. Define fitness function that takes a network as its only argument and returns fitness as float equal or greater than zero.
- If the function is written in Lisp then it must be named experiment-evaluate-network. Taking a look at xor-test.lisp, it can be seen that activate-network-with-inputs takes a network with inputs and returns a list of outputs.
- If the fitness function is written in language other than Lisp but can be accessed through C interface, it can be loaded like this (more on this later): (load-shared-object "evaluator.dll")
- Provide the starter genome off of which the whole population will be spawned. The starter genome should be minimal: the network will grow over time as needed.
Now, after loading your experiment file (like xor-test.lisp), it is possible to start the experiment with (start). There are several optional arguments used for saving and/or loading that the function start can take:
(defun start (&optional experiment-id read-generation-file) ...)
experiment-id is the id of the experiment which will frame the save-folder's name. Not supplying anything means not saving results. Supplying a number will create a folder named exp-<number> and the winner organisms will be saved there. It is necessary for saving state that the global variable save-every-generation be specified (it is in experiment-parameters.lisp).
If experiment was saved to a file, it can be read back from read-generation-file.
NOTE: experiment-id has nothing to do with read-generation-file! experiment-id is used only for saving a population. So, read-generation-file needs to contain relative path to the experiment file including all the folders (absolute path will work too).
(start nil "exp-5/gen-10.gen")
will load the generation from the specified file and will save nothing nowhere.
(start 6)
Will start a generation from scratch and will save results to folder called "exp-6" every save-every-generation times.
(start 6 "exp-5/gen-10.gen")
will load the generation from folder exp-5 and save results in "exp-6".
(start 5 "exp-5/gen-10.gen")
will load the generation from "exp-5" and save results to "exp-5" while renaming the already existing population files.
The one and only modification (as far as my vision goes) is the aging scheme, which the paper did not address. The C++ implementation by Stanley did include a variation of aging, but this package offers a different kind. The main idea is that a species is given a fitness boost depending on its age. The younger species are given boost in terms of adjusted fitness. The older the species, the less boost it gets. The organisms in the oldest species get no boost at all. The adjusted fitness value of a species for age-boosting is defined by this relationship:
where s is adjusted fitness of the species, A is the age of the oldest species, a is the age of the species and g is age significance (age-significance in the package). The age of a species is reset to zero when some organism of that species beats the best record of actual fitness inside that species.
Also, ability to cap the maximum amount of species per population is implemented. So, when a cap is reached, if some new organism is not compatible with any of the existing species in the population, instead of creating a new species, it is assigned to the one it is most compatible with.
- xor-experiment-parameters.lisp: contains all the global parameters needed for the XOR experiment.
- pln-experiment-parameters.lisp: contains all the global parameters needed for the PLN experiment.
- foreign-evaluator.lisp Uses foreign function interface to call a fitness function written in a different language and has C interface. One function should be featured in the interface:
float c_evalme(EncodedNeuron n[MAX_NEURONS_PER_NETWORK],int neuron_count);
where EncodedNeuron is a struct:
struct EncodedNeuron
{
int id;
int type; //0 - input, 1 - hidden, 2 - output
int out_nodes[MAX_NEURONS_PER_NETWORK];
float out_weights[MAX_NEURONS_PER_NETWORK];
int out_count;
};
A shared library should be provided and loaded in foreign-evaluator.lisp:
(load-shared-object "evaluator.dll")
The call to such a fitness function would be quite simple after loading:
(defun experiment-evaluate-network (network)
(call-evalme network))
- genome.lisp: defines genome and manipulation functions.
- globals.lisp: contains some global variables majorly used for constructing starter genome. Could be eliminated in the future.
- population.lisp: main file loading everything and defining population, organisms, species and start function.
- network.lisp: neural network code.
- pln-test.lisp: pln-experiment test, uses foreign function interface.
- xor-test.lisp: defines XOR experiment.
- printers.lisp: function for saving and printing info about population or organisms.
- starter.lisp: functions for constructing intitial population or reading one from file.
SBCL was the interpreter of choice.
A sample run that evolves a network to behave like a XOR-function (which requires at least one hidden node):
Notes to the picture:
- xor-test.lisp and xor-experiment-parameters.lisp were used. To run yourself, do (load xor-test.lisp) and (start).
- Population size: 150 organisms
- #sp means number of species and grows so large because compatibility threshold was set low. However, the convergence seems to be faster this way.
- When a network's output is rounded to the nearest integer and matches real XOR outputs, the fitness of the function is set to maximum fitness (sixteen) and the experiment ends.
With the parameters picked by trial and error, the average solution takes about 35 generations to evolve, which is not far from the results in the paper: 32 generations on average. The parameters could be tailored a bit better probably.
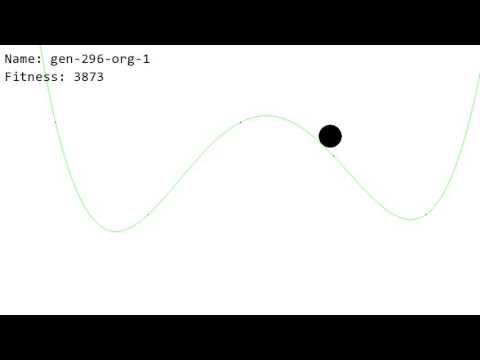
A special simulation was written for a performance test - a ball bounces on a function and the goal is to push the ball as far as possible to the right in three seconds. Here is the evolution process (when some organism sets the record for fitness, it is shown) (YouTube video):
Notes to the video:
- pln-test.lisp and pln-experiment-parameters.lisp were used. However, since the simulation is not written in Lisp, to run this experiment, you would need to compile this simulation as a shared library. Then do (load pln-test.lisp) and (start).
- Fitness starts at 2000 so that there is no way for it to be negative (though I think fitness could be negative)
- More generations would produce better results. I will post another video with those better results (and maybe even a more interesting simulation) by 2017.