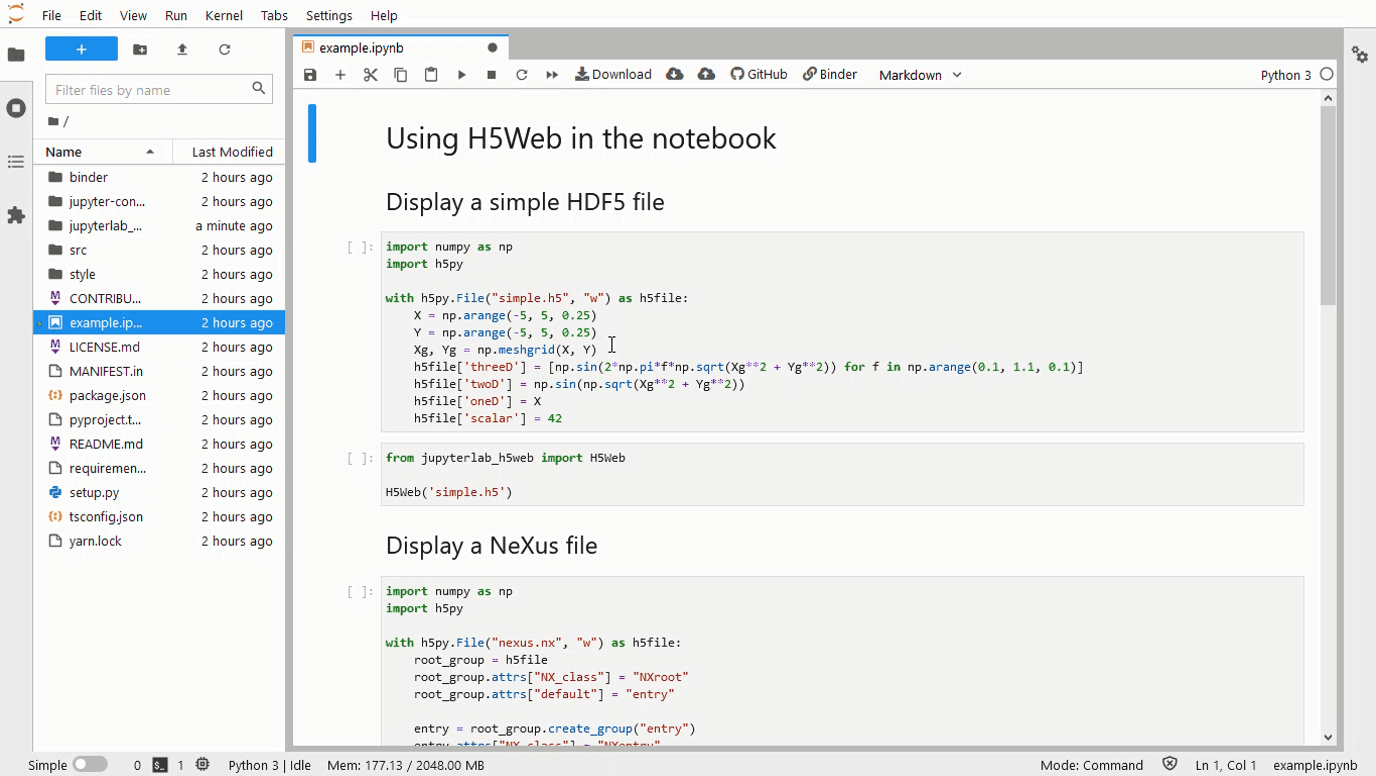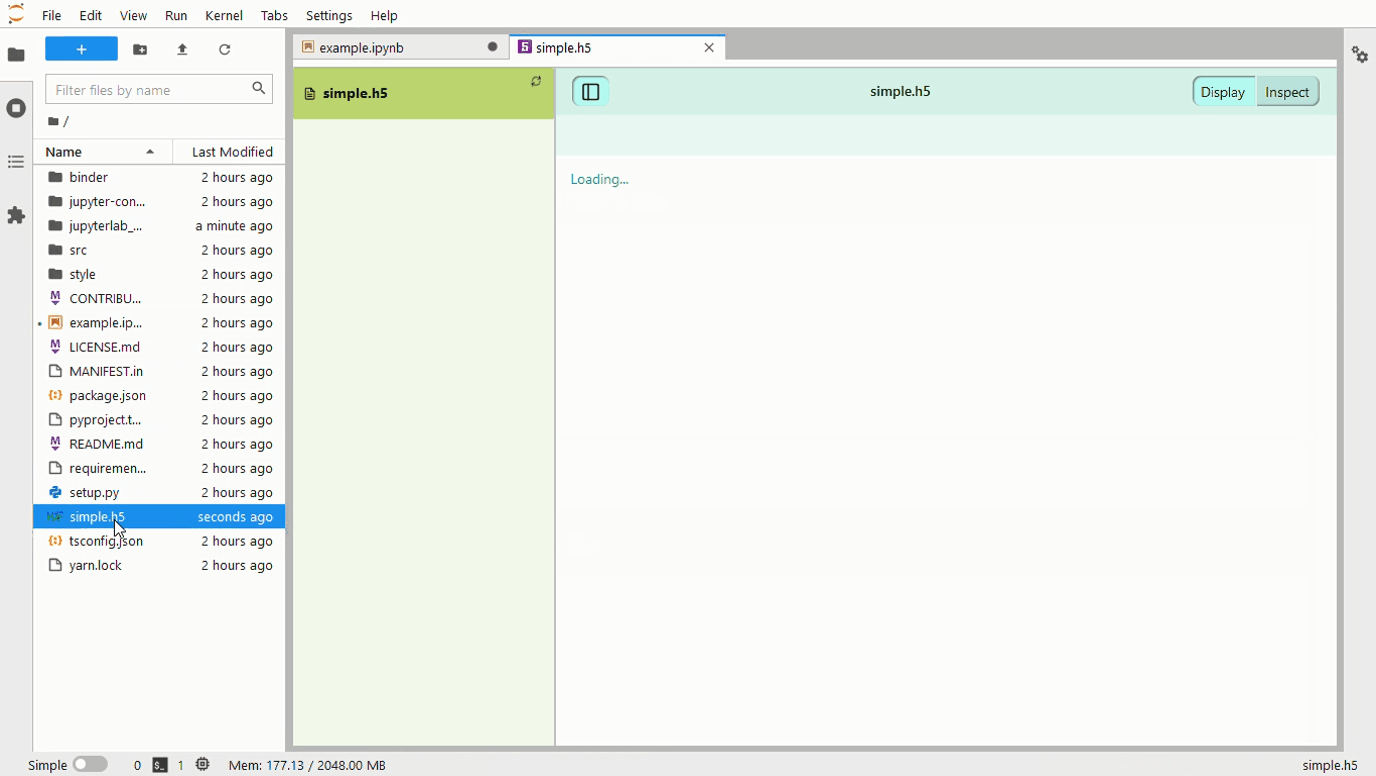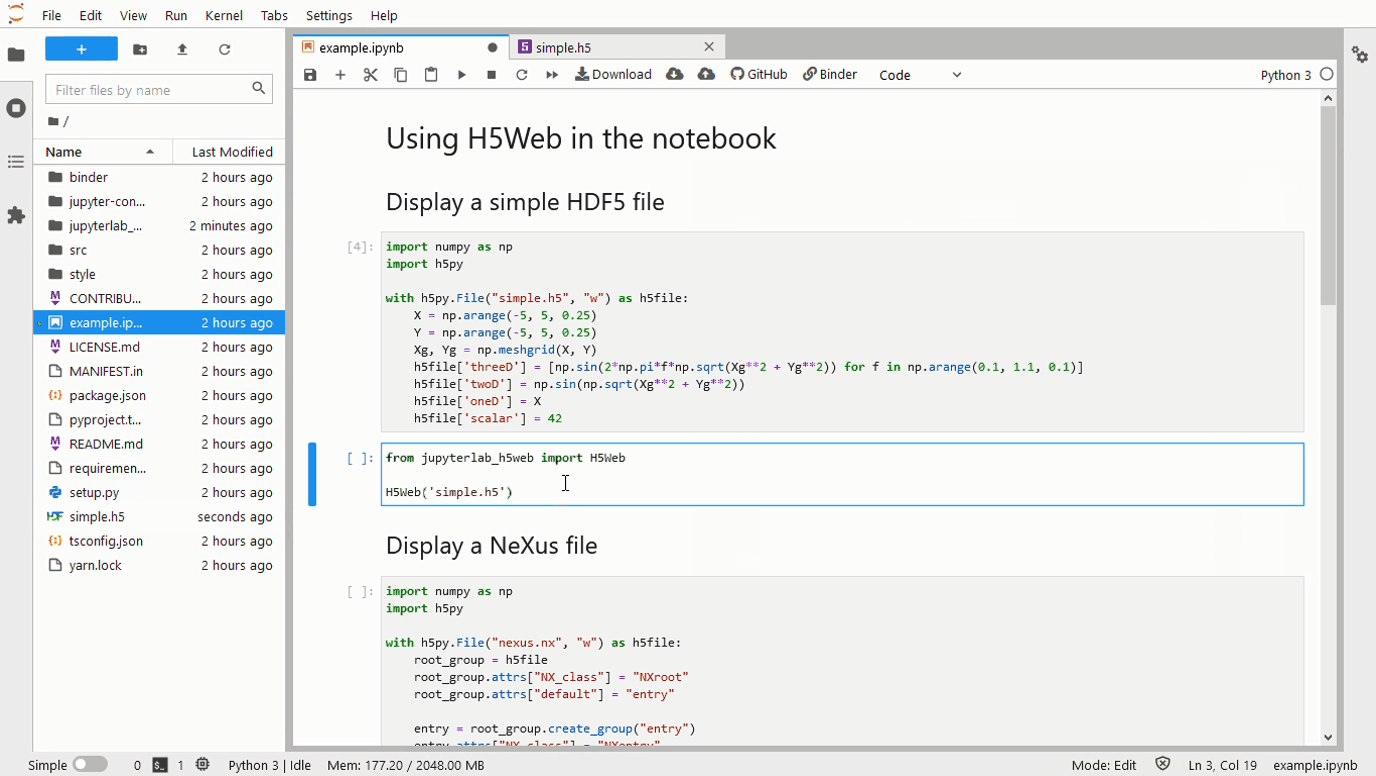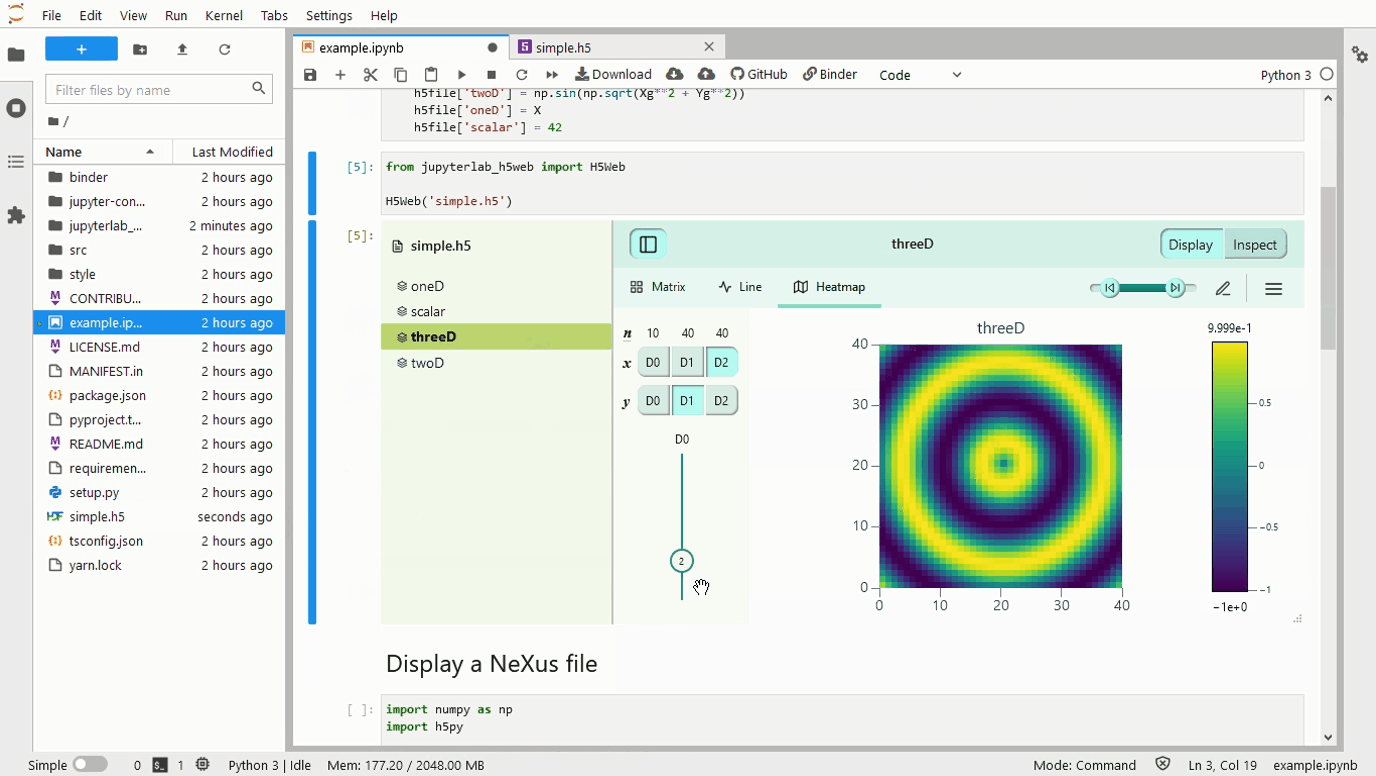
A 500 Error gets returned when a 3D array is passed to the framework. The screenshots below contain an image with a large number of the 0th dim, and another with only 67. The error log is as follows:
[E 2022-02-21 14:13:55.396 ServerApp] Uncaught exception GET /lab/api/settings?1645470835248 (127.0.0.1)
HTTPServerRequest(protocol='http', host='localhost:8888', method='GET', uri='/lab/api/settings?1645470835248', version='HTTP/1.1', remote_ip='127.0.0.1')
Traceback (most recent call last):
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/tornado/web.py", line 1704, in _execute
result = await result
tornado.iostream.StreamClosedError: Stream is closed
[I 2022-02-21 14:13:57.204 LabApp] Build is up to date
[E 2022-02-21 14:14:26.122 ServerApp] Uncaught exception GET /h5web/data/?file=MUSAN_FSD50K%2Fmusan_fsd50k_nooverlap_2s.h5&path=%2F40_mels%2Ftrain%2Ffeatures&selection=0,:,:&flatten=true (127.0.0.1)
HTTPServerRequest(protocol='http', host='localhost:8888', method='GET', uri='/h5web/data/?file=MUSAN_FSD50K%2Fmusan_fsd50k_nooverlap_2s.h5&path=%2F40_mels%2Ftrain%2Ffeatures&selection=0,:,:&flatten=true', version='HTTP/1.1', remote_ip='127.0.0.1')
Traceback (most recent call last):
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/tornado/web.py", line 1702, in _execute
result = method(*self.path_args, **self.path_kwargs)
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/tornado/web.py", line 3173, in wrapper
return method(self, *args, **kwargs)
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/jupyterlab_h5web/handlers.py", line 28, in get
response = encode(content, format_arg)
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/h5grove/encoders.py", line 109, in encode
orjson_encode(content),
File "/home/nickshu/anaconda3/envs/h5web/lib/python3.8/site-packages/h5grove/encoders.py", line 48, in orjson_encode
return orjson.dumps(content, default=default, option=orjson.OPT_SERIALIZE_NUMPY)
TypeError: default serializer exceeds recursion limit
[W 2022-02-21 14:14:26.123 ServerApp] Unhandled error
[E 2022-02-21 14:14:26.123 ServerApp] {
"Host": "localhost:8888",
"User-Agent": "Mozilla/5.0 (X11; Linux x86_64; rv:97.0) Gecko/20100101 Firefox/97.0",
"Accept": "application/json, text/plain, */*",
"Accept-Language": "en-US,en;q=0.5",
"Accept-Encoding": "gzip, deflate",
"Connection": "keep-alive",
"Referer": "http://localhost:8888/lab/workspaces/auto-x/tree/MUSAN_FSD50K",
"Cookie": "username-localhost-8888=\"2|1:0|10:1645470865|23:username-localhost-8888|44:MTBmODU5YWVjN2QwNGM3OWI0ZGIyYTg3Mzg5NDBhMDc=|71d4f38e3541a61543b4765a12ca598a1413794e587db212ad204ec9317ff226\"; _xsrf=2|be7cd264|72c37ad1b680654d763a713d31046056|1643138022",
"Sec-Fetch-Dest": "empty",
"Sec-Fetch-Mode": "cors",
"Sec-Fetch-Site": "same-origin"
}
[E 2022-02-21 14:14:26.123 ServerApp] 500 GET /h5web/data/?file=MUSAN_FSD50K%2Fmusan_fsd50k_nooverlap_2s.h5&path=%2F40_mels%2Ftrain%2Ffeatures&selection=0,:,:&flatten=true (127.0.0.1) 5.62ms referer=http://localhost:8888/lab/workspaces/auto-x/tree/MUSAN_FSD50K
Extension lists
$ jupyter labextension list
JupyterLab v3.2.9
/home/user/anaconda3/envs/h5web/share/jupyter/labextensions
jupyterlab-h5web v2.0.0 enabled OK (python, jupyterlab_h5web)
$ jupyter serverextension list
config dir: /home/user/anaconda3/envs/h5web/etc/jupyter
jupyterlab enabled
- Validating...
jupyterlab 3.2.9 OK
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 2.0.0 OK
When using Jupyter Lab's dark mode, the text in the 'inspect' view is black and almost invisible:

Similarly for the matrix view:

When I run the example for your package the object: <jupyterlab_h5web.widget.H5Web object> is displayed instead of the graphical interface- what am I doing wrong:).
H5Web allows customizing the colors in 2D plots, which is done in the css. It would be great to have a way to be able to inject this to invoking these widgets in Jupyter lab.
Originally posted by @tomio13 in #34 (comment)
Describe the bug
I am creating a dummy h5 file containing a complex array and a integer array as bellow, but I get an error message saying "Expected complex" (for the complex array) and (Expected Number) for the integer array. The same does not happen for floats; it opens normally using dtype=np.float64.
import numpy as np
import h5py
file = h5py.File('test.h5','a')
name = '1'
data = np.ones((5,5),dtype=np.complex64)
file.create_dataset(name,data=data)Screenshots

Context
- OS: Ubutun 20.04.4 LTS
- Browser: Chrome Version 112.0.5615.49 (Official Build) (x86_64)
IPython : 7.29.0
ipykernel : 6.5.0
ipywidgets : 7.6.3
jupyter_client : 7.4.9
jupyter_core : 5.2.0
jupyter_server : 1.24.0
jupyterlab : 3.6.1
nbclient : 0.5.3
nbconvert : 7.2.9
nbformat : 5.7.3
notebook : 6.4.0
qtconsole : 5.1.1
traitlets : 5.9.0
Extension lists
JupyterLab v3.6.1
/.../.local/share/jupyter/labextensions
@jupyter-widgets/jupyterlab-manager v3.1.0 enabled OK (python, jupyterlab_widgets)
/.../apps/modules/python/3.9.2/share/jupyter/labextensions
jupyterlab-plotly v5.1.0 enabled OK
jupyterlab-h5web v7.0.0 enabled OK (python, jupyterlab_h5web)
jupyter-matplotlib v0.11.2 enabled OK
config dir: /...//.local/etc/jupyter
jupyter_server_ydoc enabled
- Validating...
X is jupyter_server_ydoc importable?
config dir: /.../modules/python/3.9.2/etc/jupyter
jupyter_server_ydoc enabled
- Validating...
X is jupyter_server_ydoc importable?
jupyterlab enabled
- Validating...
jupyterlab 3.6.1 OK
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 7.0.0 OK


Console shows an Uncaught TypeError: e.current.gl is undefined
I made a recipe for jupyterlab_h5web in conda-forge: https://github.com/conda-forge/jupyterlab-h5web-feedstock
Therefore, it is installable through conda install -c conda-forge jupyterlab-h5web. It might be good to mention this in the README and other documentation. I would also be happy to add any of the developers as maintainers of the package.
Describe the bug
There seems to be a bug with the data displayed by h5web. The axis positions are not properly placed.
Running on jupyter lab 3.2.9 and jupyterlab-h5web 1.3.0.
The minimum and max values of the y axis (energy) are: −4.000e+0 to 1.000e+0
The minimum and max values of the x axis (delay) are: −2.000e+3 to 6.500e+3
To Reproduce
Expected behaviour
Screenshots

Context
- OS: ubuntu
- Browser: Chrome
- Version: 1.3.0
- JupyterLab version: 3.2.9
Extension lists
jupyter labextension list
JupyterLab v3.2.9
/home/.../jupyter/.py37env/share/jupyter/labextensions
jupyterlab-h5web v1.3.0 enabled OK (python, jupyterlab_h5web)
@jupyter-widgets/jupyterlab-manager v3.0.1 enabled OK (python, jupyterlab_widgets)
jupyter serverextension list
config dir: /home/pepe_marquez/.jupyter
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 1.3.0 OK
config dir: /home/pepe_marquez/jupyter/.py37env/etc/jupyter
jupyterlab enabled
- Validating...
jupyterlab 3.2.9 OK
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 1.3.0 OK
Describe the bug
Trying to inspect an h5 file with tree structure "/confocal/XY/...something.../raw". On initially opening in h5web under jupyter, left pane shows "/confocal" but opening this gives a blank plane.
Digging into the errors I found the following which seems to suggest the second slash is converted to '\\'
{"message": "Unhandled error", "reason": null, "traceback": "Traceback (most recent call last):\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\tornado\web.py", line 1702, in _execute\n result = method(*self.path_args, **self.path_kwargs)\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\tornado\web.py", line 3173, in wrapper\n return method(self, *args, **kwargs)\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\jupyterlab_h5web\handlers.py", line 26, in get\n content = self.get_content(h5file, path)\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\jupyterlab_h5web\handlers.py", line 59, in get_content\n return content.metadata()\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\h5grove\content.py", line 151, in metadata\n ("children", self._get_child_metadata_content(depth - 1)),\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\h5grove\content.py", line 139, in _get_child_metadata_content\n return [\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\h5grove\content.py", line 140, in \n create_content(self._h5file, os.path.join(self._path, child_path)).metadata(\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\h5grove\content.py", line 160, in create_content\n entity = get_entity_from_file(h5file, path, resolve_links)\n File "C:\Users\u1774090\Anaconda3\envs\h5web\lib\site-packages\h5grove\utils.py", line 34, in get_entity_from_file\n raise PathError(f"{path} is not a valid path in {basename(h5file.filename)}")\nh5grove.utils.PathError: /confocal\\XY is not a valid path in session-20211021-131238.hdf5\n"}
Expected behaviour
Should allow traversal of an h5 tree from root
Screenshots
Context
- OS: Windows 10
- Browser: Edge
- Version: [e.g. 22]
- H5Web context: jupyterlab_h5web + @h5web/lib
Good day,
don't know if this bug is for jyputer-h5web or for jupyter-collaboration
In jupyterlab v4 with collaborative mode got file not found because of adding "RTC:" to filepath - for example "File not found: 'RTC:discharges/2023/202306/20230629/T/1230T 2023-06-29 13:03:54.nx"
H5Web("discharges/2023/202306/20230629/T/1230T 2023-06-29 13:03:54.nx") - works
To Reproduce
Try to open hdf5/nexus file in jupyterlab with jypyter-collaboration enabled
Expected behaviour
Open file without error
Screenshots

For example csv works

but to open from python (with open(..)) "RTC:path" needs to be changed to "path")
Context
- OS: Ubuntu 22.04
- Browser: Chrome
- Version: 114
- JupyterLab version: 4.0.2
Extension lists
Paste the output from running `jupyter labextension list` and `jupyter serverextension list` from the command line here.
You may want to sanitize the paths in the output.
jupyter_lsp enabled
- Validating...
jupyter_lsp 2.2.0 OK
jupyter_nbextensions_configurator enabled
- Validating...
jupyter_nbextensions_configurator 0.6.3 OK
jupyter_server_ydoc enabled
- Validating...
X is jupyter_server_ydoc importable?
jupyterlab enabled
- Validating...
jupyterlab 4.0.2 OK
jupyterlab_git enabled
- Validating...
jupyterlab_git 0.41.0 OK
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 10.0.0 OK
nbdime enabled
- Validating...
nbdime 3.2.1 OK
jupyter-vuetify v1.8.10 enabled X
jupyterlab-h5web v10.0.0 enabled OK (python, jupyterlab_h5web)
nbdime-jupyterlab v2.2.0 enabled X
jupyterlab_pygments v0.2.2 enabled X (python, jupyterlab_pygments)
jupyter-vue v1.9.2 enabled OK
jupyter-matplotlib v0.11.3 enabled OK
@jupyter-widgets/jupyterlab-manager v5.0.7 enabled X (python, jupyterlab_widgets)
@jupyter/collaboration-extension v1.0.1 enabled OK (python, jupyter_collaboration)
@bokeh/jupyter_bokeh v3.0.7 enabled X (python, jupyter_bokeh)
@jupyterlab/git v0.41.0 enabled X (python, jupyterlab-git)
Also checked on new vm:
jupyterlab-h5web v10.0.0 enabled OK (python, jupyterlab_h5web)
jupyterlab_pygments v0.2.2 enabled X (python, jupyterlab_pygments)
@jupyter/collaboration-extension v1.0.1 enabled OK (python, jupyter_collaboration)
I am assuming this is the project behind the wonderful thing I found yesterday that lets me browse hdf5 files in jupyterlab? It looks fantastic. I wish I could figure out how to select x and y axes for a plot? I always see data versus point number. The rest of the message is a bug report for how I seem to have broke something already (sorry!) :
Describe the bug
jupyterlab crashes when reading large dataset, perhaps an out of memory error?
To Reproduce
1 - Log into jupyter-slurm.esrf.fr with one single core and the lab interface
2 - Navigate to open : /data/id11/nanoscope/blc12407/id11/CeO2_38keV/CeO2_38keV_CeO2_rotation/CeO2_38keV_CeO2_rotation.h5
3 - open dataset /1.1/measurement/eiger : it displays
4 - open dataset /1.1/measurement/fpico6 : it displays
5 - go back to /1.1/measurement/eiger : jupyterlab stops running
6 - all the other tabs and kernels appear to exit when jupyterlab fails
Expected behaviour
In the worst case, a plugin would crash without taking down all of the other kernels. Ideally it would not crash.
Is there a way to use hdf5 slice operations (maybe combined with fast histograms) so you only hold in memory what is going to be displayed on the screen (e.g. maximum data is a 2D image)? Then libhdf5 should manage the memory cache in some sensible way.
Context
- OS: ubuntu20.04
- Browser: Chrome
- Version: 94.0.4606.81
- JupyterLab version: 2.3.1 (ESRF slurm installation
Extension lists
This is based on a bit of guesswork as to what is actually running when I use jupyter-slurm :
jupyter-slurm:~ % /scisoft/users/jupyter/jupy38ubuntu/bin/jupyter labextension list
JupyterLab v2.3.1
Known labextensions:
app dir: /home/esrf/jupyter/jupy38ubuntu/share/jupyter/lab
@jupyter-widgets/jupyterlab-manager v2.0.0 enabled OK
jupyter-matplotlib v0.7.4 enabled OK
jupyter-threejs v2.2.0 enabled OK
jupyterlab-datawidgets v6.3.0 enabled OK
jupyterlab-h5web v0.0.10 enabled OK
k3d v2.9.3 enabled OK
jupyter-slurm:~ % /scisoft/users/jupyter/jupy38ubuntu/bin/jupyter serverextension list
config dir: /home/esrf/jupyter/jupy38ubuntu/etc/jupyter
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web OK
jupyterlab enabled
- Validating...
jupyterlab 2.3.1 OK
jupyterlab_hdf enabled
- Validating...
jupyterlab_hdf 0.5.1 OK
jupyter_nbextensions_configurator enabled
- Validating...
jupyter_nbextensions_configurator 0.4.1 OK
This would be inline with h5py and h5glance behaviour.
Describe the bug
When the extension was installed in "dev mode", it is not possible to display a R3F visualisation: it freezes the browser.
Displaying scalar values, navigating in the file or inspecting entities do not trigger this freeze.
To Reproduce
- Install the extension according to https://github.com/silx-kit/jupyterlab-h5web/blob/main/CONTRIBUTING.md#install
- Run
jupyter lab
- Open a HDF file with nD datasets inside
- Open a nD dataset
- The browser will freeze
Interestingly, this does not happen with a regular installation pip install .
Context
- Browser: any
- Version: 1.0.0
- JupyterLab version: 3
Reported in #48
Dear h5web-Team,
I created a h5 file with a virtual dataset. I am following, in general, this example: https://github.com/h5py/h5py/blob/master/examples/eiger_use_case.py
As a result, I get a h5 file, where data contains my 3D stack in '/data/'.
I thought I check the content out in h5web because I work on a jupyterhub. I get "Request failed with status code 500"
What is the minimum requirement to visualize your own created h5 files with h5web, please?
In silx, I am able to see my data.
Thank you.
Hi, I was using jupyterlab-h5web on jupyter-slurm.esrf.fr, and some files are veeeery slow, and I have one that doesn't even open. I am not sure if this is somehow expected/normal, but I recorded a few of this issues so you can check it.
See: https://youtu.be/qSM2JsVVEHY
I open 4 files in the video, respectively:
-
0:08: an .h5 from the acquisition system at id11
path: /data/id11/3dxrd/blc12852/id11/bmg_l1/bmg_l1_bmg_dct2/bmg_l1_bmg_dct2.h5
issue: 0:38: displaying one frame is quite slow
-
1:15: a "small" .h5 written by my code is quite slow
path: /data/id11/3dxrd/blc12852/id11/analysis/bmg_l1_bmg_dct2/difspots/000000.h5
issue: isn't it slow relative to the size of the things inside?
-
1:33: an .h5 regrouping (external links) about 3300 .h5s like the one above
path: /data/id11/3dxrd/blc12852/id11/analysis/bmg_l1_bmg_dct2/difspots.h5
issue: slow to open, then very slow to expand the group with the external links
-
2:30: an .h5 with only 2 datasets, about the same size of the first one
path: /data/id11/3dxrd/blc12852/id11/bmg_l1/bmg_l1_bmg_dct2/bmg_l1_bmg_dct2.preprocessed.h5
issue: veeeeeeeery slow; I kept waiting after the end of the video for several minutes, the entire jupyterlab becomes very slow, and it ends up breaking the page (screenshot below). When I reload the page, the h5web tab (in jupyterlab) is not there anymore.

A few notes (Idk if this might have some influence):
- I am using symbolic links (
ln -s) to be able to see those files in jupyter-slurm because they are in /data.
- I am writting my
.h5 files with silx.io.dictdump.dicttoh5. I can show you specifically where in my code.
- I saw similar effects on a
jupyterlab instances on gpid11-nice and rnice8 nodes. In these cases I launch jupyterlab on those machines, connect to them with ssh -L 8888:localhost:8888 to forward the application's port, then access it using my local browser (my personal computer, not an ESRF one).
Running H5Web("new_file+d.h5") from a notebook leads to opening "new_file d.h5" on the server side (its already changed in the GET request).
Logs on the server side:
Traceback (most recent call last):
File "lib/python3.8/site-packages/tornado/web.py", line 1702, in _execute
result = method(*self.path_args, **self.path_kwargs)
File "lib/python3.8/site-packages/tornado/web.py", line 3173, in wrapper
return method(self, *args, **kwargs)
File "lib/python3.8/site-packages/jupyterlab_h5web/handlers.py", line 25, in get
with h5py.File(as_absolute_path(self.base_dir, Path(file_path)), "r") as h5file:
File "lib/python3.8/site-packages/h5py/_hl/files.py", line 424, in __init__
fid = make_fid(name, mode, userblock_size,
File "lib/python3.8/site-packages/h5py/_hl/files.py", line 190, in make_fid
fid = h5f.open(name, flags, fapl=fapl)
File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper
File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper
File "h5py/h5f.pyx", line 96, in h5py.h5f.open
OSError: Unable to open file (unable to open file: name = '/home/new_file d.h5', errno = 2, error message = 'No such file or directory', flags = 0, o_flags = 0)
The display of 2D datasets makes the viewer crash 🙁
Installation fails with:
ERROR: Could not build wheels for jupyterlab-h5web which use PEP 517 and cannot be installed directly
While a potential workaround could have been:
pip install --no-use-pep517 jupyterlab_h5web
this also fails with:
ERROR: Disabling PEP 517 processing is invalid: project specifies a build backend of
setuptools.build_meta in pyproject.toml
Ideas? Thanks!
We are typically working with files that have a different extension than .h5 or .hdf5. Those can only be opened after renaming them as far as I can tell. Even the 'Open with' menu lists only the editor, not h5web.
It is probably not useful to list all custom extensions that people use. But it would be great if the viewer would not be tied to the file name.
I've been trying to install the extension on ubuntu 20.04 and I'm getting the following conflicting dependency error message
[LabBuildApp] JupyterLab 2.2.9
[LabBuildApp] Building in $HOME/anaconda3/envs/hdf5/share/jupyter/lab
[LabBuildApp] Building jupyterlab assets (build:prod:minimize)
[LabBuildApp] WARNING |
"[email protected]" is not compatible with the current JupyterLab
Conflicting Dependencies:
JupyterLab Extension Package
>=2.2.6 <2.3.0 >=2.3.0 <2.4.0||>=3.0.5 <4.0.0@jupyterlab/apputils
>=2.2.4 <2.3.0 >=2.3.0 <2.4.0||>=3.0.7 <4.0.0@jupyterlab/rendermime
>=2.2.1 <2.3.0 >=2.3.0 <2.4.0||>=3.0.6 <4.0.0@jupyterlab/rendermime-interfaces
JupyterLab v2.2.9
Known labextensions:
app dir: $HOME/anaconda3/envs/hdf5/share/jupyter/lab
@jupyterlab/hdf5 v0.5.1 enabled OK
jupyterlab-h5web v0.0.7 enabled XThanks!
Toolbar controls are shared among JupyterLab tabs, even if they display different files.
Hello!
My group is interested to explore an H5 dataset through plotting various dimensions across each other. Your JupyterLab plugin interface is awesome, but we cannot seem select our own dimensions to plot. Can your plugin accommodate selecting data dimensions for plotting, or can the code be expanded to add this functionality?
Andrew
See the discussion at #12
Good day
Looks like that legend color changes with theme switch, but not the lines color. (Incorrect view in dark mode)
Screenshots


Context
- OS: Ubuntu 22.04.2 LTS
- Browser: Chrome
- Version: 7.1.2
- JupyterLab version: 2.5.0
Extension lists
jupyterlab-h5web v7.1.2 enabled OK (python, jupyterlab_h5web)
jupyterlab_pygments v0.2.2 enabled OK (python, jupyterlab_pygments)
jupyter_server_ydoc enabled
- Validating...
X is jupyter_server_ydoc importable?
jupyterlab enabled
- Validating...
jupyterlab 3.6.3 OK
jupyterlab_h5web enabled
- Validating...
jupyterlab_h5web 7.1.2 OK
The extension cannot be enabled with Jupyter Lab 3.1.x
Expected Result:
$ jupyter labextension list
JupyterLab v3.1.1
Other labextensions (built into JupyterLab)
app dir: ~/.venv/share/jupyter/lab
jupyterlab-h5web v0.0.8 enabled **OK**
Actual Result
$ jupyter labextension list
JupyterLab v3.1.1
Other labextensions (built into JupyterLab)
app dir: ~/.venv/share/jupyter/lab
jupyterlab-h5web v0.0.8 enabled **X**
The following extension are outdated:
jupyterlab-h5web
Workaround
I think the last version with which it could be enabled was Jupyter Lab 3.0.16
When opening a hdf5 file, the widget briefly (milliseconds) pops-up, to close again immediately.
Other widgets and interactive elements have no problems.
I just used the sample notebook to replicate the issue.
Tried multiple browsers: FF, Edge, Chrome on Win10.
Tried versions v0.0.10, 9 and 8.
Using jupyterlab v3.0.16
Settings > JupyterLab Theme > JupyterLab Light

All is well.
Settings > JupyterLab Theme > JupyterLab Dark

Heatmap, color map and color bar are inverted.
In its current implementation, the extension basically merges H5Web's CSS and JS code into Jupyer Lab's:
- H5Web's global styles (
normalize.css and .btn-clean, variables, etc.) are merged with Jupyter Lab's styles, which leads to unexpected results - e.g. H5Web's built-in support for system dark mode (prefers-color-scheme: dark) applies filter: invert() to JupyterLab as a whole, even when JLab is configured with a light theme.
- H5Web's JS bundle is executed on launch of JLab, which means any global state is shared between JLab tabs. This is what led us to switching from global zustand stores to context-injected stores.
All of these issues, and likely future issues, would be avoided if every instance of H5Web (tab or cell) were completely self-contained. The easiest way to achieve JS/CSS containment is to use an iframe.
The way I see it, the extension could take the form of a little create-react-app that renders H5Web's App component (wrapped in a JupyterProvider) on a simple HTML page. Building the extension would generate all the static assets (index.html + JS/CSS bundles). On JLab start-up, the extension would start a static file server (or use JLab's) to serve those static assets at https://jlab.url/h5web/?file=some-file.h5. When opening an HDF5 file in a JLab tab or cell, the extension would then render an iframe with its src attribute set to that URL.
I have no idea how feasible this solution is...
Is your feature request related to a problem? Please describe.
I think this plugin can handle netcdf files as they are CF compliant HDF files, I tested changing the file extension to some .nc files and it worked fine.
Describe the solution you'd like A clear and concise description of what you
want to happen.
Would it be possible to include netcdf in the list of supported formats?
Describe alternatives you've considered A clear and concise description of
any alternative solutions or features you've considered.
Additional context Add any other context or screenshots about the feature
request here.
When the top-level HDF5 groups are external links (BLISS proposal and sample collection files), the entire file doesn't show up and this traceback is printed:
[E 2021-06-16 23:54:31.907 ServerApp] Found and opened file, error getting contents from object specified by the uri.
Error: Traceback (most recent call last):
File "/users/denolf/virtualenvs/jupyter/ubuntu_20_04/lib/python3.9/site-packages/jupyterlab_hdf/baseHandler.py", line 72, in get
out = self._get(fpath, uri, **kwargs)
File "/users/denolf/virtualenvs/jupyter/ubuntu_20_04/lib/python3.9/site-packages/jupyterlab_hdf/baseHandler.py", line 90, in _get
return self._getFromFile(f, uri, **kwargs)
File "/users/denolf/virtualenvs/jupyter/ubuntu_20_04/lib/python3.9/site-packages/jupyterlab_hdf/meta.py", line 16, in _getFromFile
return jsonize(hobjMetaDict(f[uri], ixstr=ixstr, min_ndim=min_ndim))
File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper
File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper
File "/users/denolf/virtualenvs/jupyter/ubuntu_20_04/lib/python3.9/site-packages/h5py/_hl/group.py", line 288, in __getitem__
oid = h5o.open(self.id, self._e(name), lapl=self._lapl)
File "h5py/_objects.pyx", line 54, in h5py._objects.with_phil.wrapper
File "h5py/_objects.pyx", line 55, in h5py._objects.with_phil.wrapper
File "h5py/h5o.pyx", line 190, in h5py.h5o.open
KeyError: "Unable to open object (object '8.1' doesn't exist)"
relfpath: data/inhouse/id002106/id00/id002106_id00.h5, uri: /8.1, min_ndim: None, ixstr: None, subixstr: NoneThe HDF5 group of the link destination is indeed /8.1 but the relfpath is the path of the file that contains the link, not the path of the file of the link destination.
>>> import os, h5py
>>> os.getcwd()
'/tmp/data/inhouse/id002106/id00'
>>> f = h5py.File("id002106_id00.h5")
>>> f.filename
'id002106_id00.h5'
>>> f["sample_0005_8.1"].file.filename
'/tmp/data/inhouse/id002106/id00/sample/sample_0005/sample_0005.h5'
>>> f["sample_0005_8.1"].name
'/8.1'
>>> link = f.get("sample_0005_8.1", getlink=True)
>>> link.filename
'sample/sample_0005/sample_0005.h5'
>>> link.path
'/8.1'The jupyterlab working directory was /tmp
Recommend Projects
-
-
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
An Open Source Machine Learning Framework for Everyone
-
The Web framework for perfectionists with deadlines.
-
A PHP framework for web artisans
-
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
Some thing interesting about web. New door for the world.
-
A server is a program made to process requests and deliver data to clients.
-
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Some thing interesting about visualization, use data art
-
Some thing interesting about game, make everyone happy.
-
Recommend Org
-
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Open source projects and samples from Microsoft.
-
Google ❤️ Open Source for everyone.
-
Alibaba Open Source for everyone
-
Data-Driven Documents codes.
-
China tencent open source team.
-