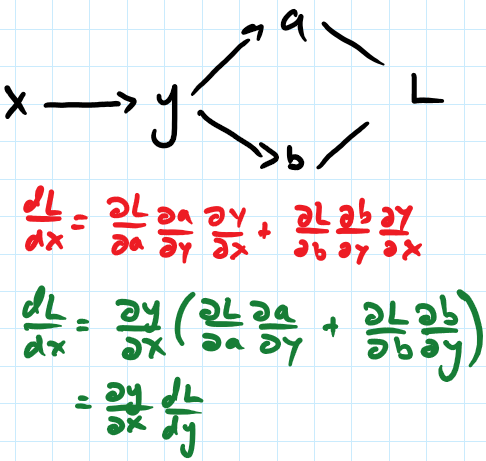
MyGrad is a lightweight library that adds automatic differentiation to NumPy – its only dependency is NumPy! It is specifically able to compute gradients of scalar-valued functions via backpropagation (i.e. reverse-mode automatic differentiation).
>>> import mygrad as mg
>>> import numpy as np
>>> x = mg.tensor([1., 2., 3.]) # like numpy.array, but supports backprop!
>>> f = np.sum(x * x) # tensors work with numpy functions!
>>> f.backward() # triggers automatic differentiation
>>> x.grad # stores [df/dx0, df/dx1, df/dx2]
array([2., 4., 6.])MyGrad's primary goal is to make automatic differentiation accessible and easy to use across the Python/NumPy ecosystem. As such, it strives to behave and feel exactly like NumPy so that users need not learn yet another array-based math library. Of the various modes and flavors of auto-diff, MyGrad supports backpropagation from a scalar quantity.
Installing MyGrad:
pip install mygradNumPy's ufuncs are richly supported; e.g. we can autodiff through in-place targets and boolean masks:
>>> x = mg.tensor([1., 2., 3.])
>>> y = mg.zeros_like(x)
>>> np.multiply(x, x, where=[True, False, True], out=y)
>>> y.backward()
>>> x.grad
array([2., 0., 6.])NumPy's view semantics are also mirrored to a high fidelity
>>> x = mg.arange(9.).reshape(3, 3)
>>> diag_view = np.einsum("ii->i", x)
>>> x, diag_view
(Tensor([[0., 1., 2.],
[3., 4., 5.],
[6., 7., 8.]]),
Tensor([0., 4., 8.]))
# views share memory
>>> np.shares_memory(x, diag_view)
True
# mutating a view affects its base (and all other views)
>>> diag_view *= -1 # mutates x in-place
>>> x
Tensor([[-0., 1., 2.],
[ 3., -4., 5.],
[ 6., 7., -8.]])
>>> (x ** 2).backward()
>>> x.grad, diag_view.grad
(array([[ -0., 2., 4.],
[ 6., -8., 10.],
[ 12., 14., -16.]]),
array([ -0., -8., -16.]))
# the gradients have the same view relationship!
>>> np.shares_memory(x.grad, diag_view.grad)
TrueBasic and advanced indexing is fully supported
>>> (x[x < 4] ** 2).backward()
>>> x.grad
array([[0., 2., 4.],
[6., 0., 0.],
[0., 0., 0.]])NumPy arrays and other array-likes play nicely with MyGrad's tensor. These behave like constants during automatic differentiation
>>> x = mg.tensor([1., 2., 3.])
>>> y = np.array([-1., 0., 10])
>>> (x * y).backward() # y is treated as a constant
>>> x.grad
array([-1., 0., 10.])mygrad.nnet supplies essential functions to facilitate typical machine learning examples:
- N-dimensional convolutions (with striding, dilation, and padding)
- N-dimensional pooling
- A gated recurrent unit for sequence-learning (with input-level dropout and variational hidden-hidden dropout)
The following is an example of using mygrad to compute the hinge loss of classification scores and to "backpropagate" through (compute the gradient of) this loss. This example demonstrates some of mygrad's ability to perform backpropagation through broadcasted operations, basic indexing, advanced indexing, and in-place assignments.
>>> import mygrad as mg
>>> import numpy as np
>>> class_scores = 10 * mg.random.rand(100, 10) # 100 samples, 10 possible classes for each
>>> class_labels = np.random.randint(low=0, high=10, size=100) # correct label for each datum
>>> class_labels = (range(len(class_labels)), class_labels)
>>> correct_class_scores = class_scores[class_labels]
>>> Lij = class_scores - correct_class_scores[:, np.newaxis] + 1. # 100x10 margins
>>> Lij[Lij <= 0] = 0 # scores within the hinge incur no loss
>>> Lij[class_labels] = 0 # the score corresponding to the correct label incurs no loss
>>> loss = Lij.sum() / class_scores.shape[0] # compute mean hinge loss
>>> loss.backward() # compute gradient of loss w.r.t all dependent tensors
>>> class_scores.grad # d(loss)/d(class_scores)
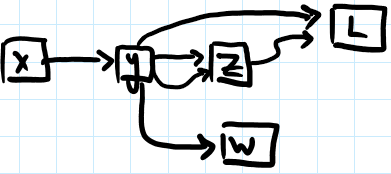
array([[ 0. , 0.01, 0. , -0.04, 0. , 0. , 0.01, 0. , 0.01, 0.01], ...])mygrad uses Graphviz and a Python interface for Graphviz to render the computational graphs built using tensors. These graphs can be rendered in Jupyter notebooks, allowing for quick checks of graph structure, or can be saved to file for later reference.
The dependencies can be installed with:
conda install graphviz
conda install python-graphviz