Online RADIS environment for advanced processing and comparison with experimental spectra
Online environment for advanced spectrum processing and comparison with experimental data
Home Page: https://radis.github.io/radis-lab
Online RADIS environment for advanced processing and comparison with experimental spectra

Add custom parameters for Radis-lab by default :
See https://github.com/ian-r-rose/binder-workspace-demo :
The easiest way to make a workspace is to launch JupyterLab and arrange the application in the layout you prefer. Once you have your layout, open a terminal and enter
mkdir binder
jupyter lab workspaces export > binder/workspace.json
Radis-Lab links can already be shared (distant user access the latest saved version)
but adding the --collaborative option should allow real-time collaboration :
Also :
see jupyterlab/jupyterlab#11304
to be able to see ~/.radisdb files
In the latest commits to RADIS-Lab 567b025 I added the auto-download of HITEMP H2O lines.
Challenge is to be able to run a large range, high-temperature H2O computation directly on RADIS-Lab ! (no install needed !) 🥇 🥇
Currently, we are limited because :
Fix :
Apps and Notebooks related to RADIS that could be useful !
Basically something along the line of :

on a Plotly/Streamlit/Holoview interactive dashboard, along one to choose molecules, & a slider for filter center and HWHM ?
New Dashboard for Fitroom. Currently a private repo (ask for access on the Slack !)
Tip : you can vote for New RADIS Features on https://feathub.com/radis/radis
Show examples under a nice HTML Gallery ?
An .html could be generated and shown on 🌱 RADIS-lab start.
Users would start with a page similar to this: https://sphinx-gallery.github.io/stable/auto_examples/index.html#gallery-of-examples
Ressources :
(linked to the same need for the main RADIS repo radis/radis#136)
Implement this :
https://github.com/marketplace/actions/trigger-binder-build
Note :
Since binder is an none profit project, please use this action sparsely and in a none spammy manner, to treat their resources with care.
👉 let's only use on main branch.
I'm not used to Github Actions, but I think we can use this :
name: 'Trigger-Binder-build'
on:
push:
branches:
- main
jobs:
trigger-binder-build:
runs-on: [ubuntu-latest]
steps:
- uses: s-weigand/trigger-mybinder-build@v1
with:
target-repo: radis/radis-lab
Not directly related : afterwards we'll probably also use GitHub Actions to build a part of the docker and reduce the charge on Binder. See #13
Using %matplotlib widget makes plot interactive but they are not exported in notebooks with nbconvert (or nbviewer .
As long as it is not fixed we could default to %matplotlib inline and add a comment to let users change it to %matplotlib widget if they want interaction.
Steps to reproduce :
https://nbviewer.jupyter.org/github/radis/radis-lab/blob/main/welcome.ipynb
As of 02/01/21 : shows a 404 error (but the old "welcome.md" is found)
Adding ?flush_cache=true does not work. jupyter/nbviewer#972
(nbviewer is used while loading Binder)
See https://jupyterlab.readthedocs.io/en/stable/user/files.html
For the parsing, one can use hit2df
We would need two repo : a new "radis-lab-env" repo with Anaconda & the databases (unzipped and cached), which generates a first Docker, and this one "radis-lab" with the notebooks that builds on the first Docker, so we can modify them without having to rebuild everything
See how : https://discourse.jupyter.org/t/how-to-reduce-mybinder-org-repository-startup-time/4956
This would also allow to use the "radis-lab-env" repo for other Forks, e.g. a student class with only the required notebooks.
Implementation :
we can have GitHub Actions build the Dockers. See link above, and also : https://github.com/jupyterhub/repo2docker-action
Note : if so then we'll have to remove the Binder Build Action #16
A first attempt to run RADIS on Google-colab with GPU (note that GPU must be enabled in Edit / Notebook settings)
https://colab.research.google.com/drive/1RnbAm_YIBZJW-7fEXp60oGFX8iAt_951#scrollTo=WRHD7xue_-yw
!pip install radis
Got radis 0.10.3
from radis import calc_spectrum
s = calc_spectrum(1900, 2300, # cm-1
molecule='CO',
isotope='1,2,3',
pressure=1.01325, # bar
Tgas=700, # K
mole_fraction=0.1,
path_length=1, # cm
databank='hitemp', # or use 'hitemp'
verbose=False,
mode='gpu'
)
s.apply_slit(0.5, 'nm') # simulate an experimental slit
s.plot('radiance')Failed with the following :
RADIS configuration file created in /root/radis.json
Added HITEMP-CO database in /root/radis.json
---------------------------------------------------------------------------
FileNotFoundError Traceback (most recent call last)
<ipython-input-3-428ebed103f5> in <module>()
9 databank='hitemp', # or use 'hitemp'
10 verbose=False,
---> 11 mode='gpu'
12 )
13 s.apply_slit(0.5, 'nm') # simulate an experimental slit
3 frames
/usr/local/lib/python3.7/dist-packages/radis/lbl/gpu.py in <module>()
67 # TO-DO: read and compile CUDA code at install time, then pickle the cuda object
68 cuda_fname = join(getProjectRoot(), "lbl", "gpu.cu")
---> 69 with open(cuda_fname, "rb") as f:
70 cuda_code = f.read().decode()
71
FileNotFoundError: [Errno 2] No such file or directory: '/usr/local/lib/python3.7/dist-packages/radis/lbl/gpu.cu'
postBuild. Ex :jupyter nbconvert --to notebook --execute compare_with_experiment.ipynb
Note : nbconvert wdoes not have an option --to none not to write an output file . jupyter/nbclient#4 . The alternative is to use papermill
papermill {test_file_name.ipynb} /dev/null
And then
Instead no file is opened on launch, despite launch adress being https://mybinder.org/v2/gh/radis/radis-lab/main?urlpath=lab/tree/welcome.ipynb
I think this started with 968970e
Previously working : #2
Investigating JupyterLab workspaces .
Seen during launch of #33
65536 bytes written to /home/jovyan/.radisdb/hitran/downloads__can_be_deleted/H2O/H2O_7.data
65536 bytes written to /home/jovyan/.radisdb/hitran/downloads__can_be_deleted/H2O/H2O_7.data
65536 bytes written to /home/jovyan/.radisdb/hitran/downloads__can_be_deleted/H2O/H2O_7.data
65536 bytes written to /home/jovyan/.radisdb/hitran/downloads__can_be_deleted/H2O/H2O_7.data
Header written to /home/jovyan/.radisdb/hitran/downloads__can_be_deleted/H2O/H2O_7.header
END DOWNLOAD
Lines parsed: 23192
PROCESSED
PROCESSED
Traceback (most recent call last):
File "download_hitran.py", line 10, in <module>
fetch_hitran(molecule, parse_quanta=molecule in ['CO', 'CO2'])
File "/srv/conda/envs/notebook/lib/python3.8/site-packages/radis/io/hitran.py", line 1407, in fetch_hitran
ldb.download_and_parse(download_files, cache=cache, parse_quanta=parse_quanta)
File "/srv/conda/envs/notebook/lib/python3.8/site-packages/radis/io/hitran.py", line 1179, in download_and_parse
df = hit2df(
File "/srv/conda/envs/notebook/lib/python3.8/site-packages/radis/io/hitran.py", line 208, in hit2df
mol = get_molecule(int(f.read(2)))
ValueError: invalid literal for int() with base 10: ''
Warning : 3 files were written but not combined. Deleting them.
['/home/jovyan/.radisdb/hitran/H2O_temp000000.hdf5', '/home/jovyan/.radisdb/hitran/H2O_temp000001.hdf5', '/home/jovyan/.radisdb/hitran/H2O_temp000002.hdf5']
Full log in #33 (comment)
https://notebooks.gesis.org/hub/binder/v2/gh/radis/radis-lab/main?urlpath=lab/tree/welcome.ipynb doesn't work anymore
The whole https://notebooks.gesis.org/hub/binder/ service seems down.
Directly from this repo, for individual use on personal laptops.
Use :
jupyter lab welcome.ipynb ?Small enhancements to be worked on + not worth opening an issue. Suggest in comments !
instead of mybinder.org.
(note : will use this Fix to try the Github Action autobuild #16 )
JupyterLab HDF5 extension was added in 66a0149
Issues opening the cache file generated for the HITEMP CO lines :
No access to the file metadata ?
block0_values : no column names. Is that normal ?
Currently not possible with JupyterLab : jupyterlab/jupyterlab#2405
But it may be doable at runtime : https://discourse.jupyter.org/t/changing-favicon-with-notebook-extension/2721
We could use one of :
PS : for future reference. How to switch between the two logo for dark mode https://dev.to/lauragift21/how-to-switch-logo-in-dark-mode-2355
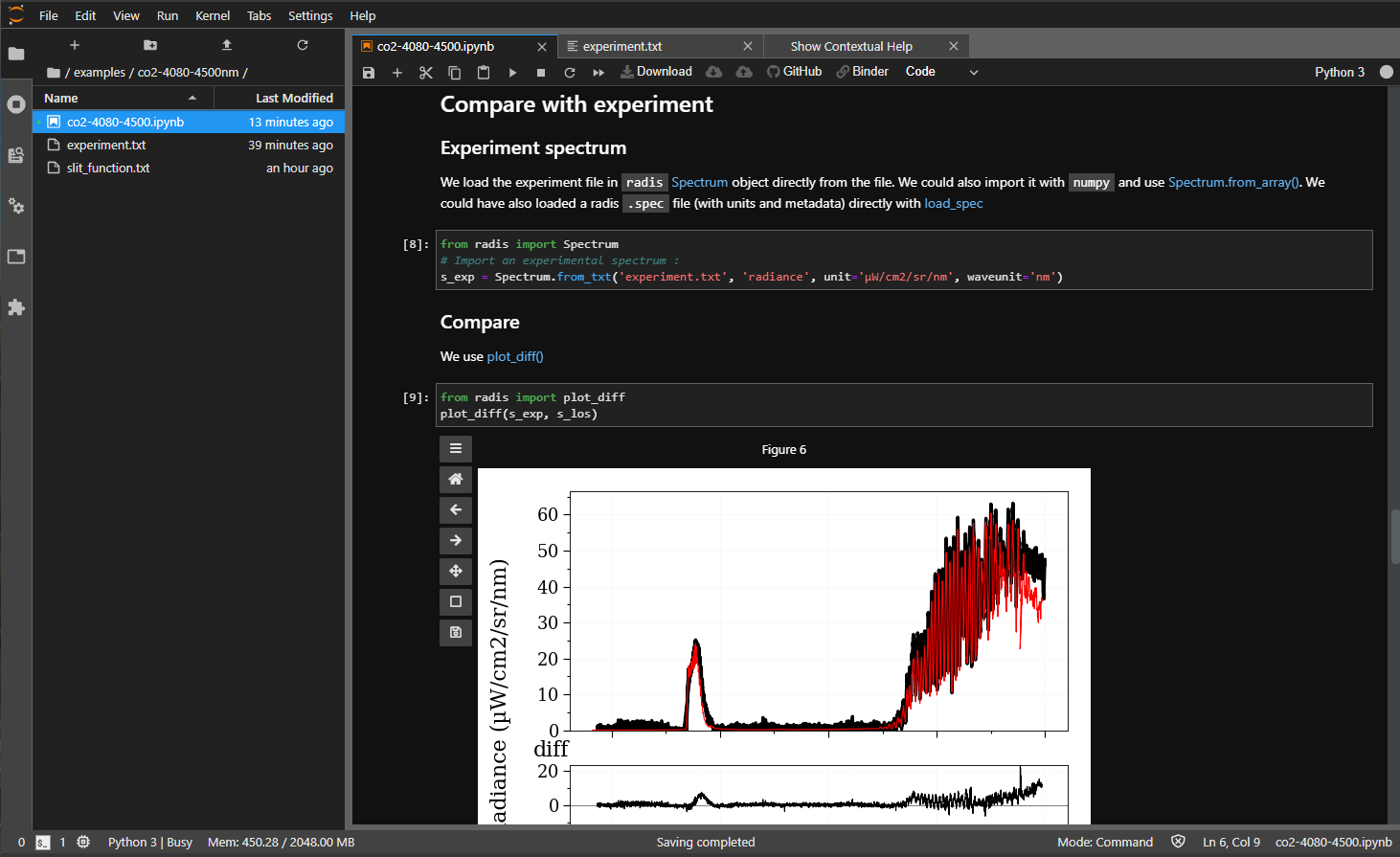
In this example of Comparing line-of-sight CO2 with experiments, executed by a https://notebooks.gesis.org/ binder on radis-lab, when running this cell:
from radis.test.tools.test_slit import linear_dispersion
dispersion = lambda w: linear_dispersion(w, 756, phi=-6.91, m=1, gr=297.42) # Reciprocal linear dispersion of the Acton 750i
s_los.apply_slit("slit_function.txt", slit_dispersion=dispersion)An AssertionError appears:
---------------------------------------------------------------------------
AssertionError Traceback (most recent call last)
/tmp/ipykernel_735/2481538297.py in <module>
1 from radis.test.tools.test_slit import linear_dispersion
2 dispersion = lambda w: linear_dispersion(w, 756, phi=-6.91, m=1, gr=297.42) # Reciprocal linear dispersion of the Acton 750i
----> 3 s_los.apply_slit("slit_function.txt", slit_dispersion=dispersion)
/srv/conda/envs/notebook/lib/python3.8/site-packages/radis/spectrum/spectrum.py in apply_slit(self, slit_function, unit, shape, center_wavespace, norm_by, mode, plot_slit, store, slit_dispersion, slit_dispersion_threshold, auto_recenter_crop, verbose, inplace, *args, **kwargs)
2729 self._q["wavespace"], w_conv
2730 ):
-> 2731 raise AssertionError(
2732 "Wavespace of convolved arrays is different, cannot store it in the same Spectrum. You can use Spectrum.apply_slit(inplace=False) to return a new spectrum with only the convolved arrays"
2733 )
AssertionError: Wavespace of convolved arrays is different, cannot store it in the same Spectrum. You can use Spectrum.apply_slit(inplace=False) to return a new spectrum with only the convolved arrays
It is worth mentioning that same AssertiveError message pops up when this cell, also uses apply_slit(), is being run:
s.apply_slit("slit_function.txt", slit_dispersion=dispersion)
s.plot(wunit='nm')
s_los.plot(wunit='nm', nfig='same')
#import matplotlib.pyplot as plt
#plt.xlim((4165, 4180))Currently, adding argument inplace=False into the function seems to prevent the error from showing up and let the function run normally.
See the following jupyterlab/jupyterlab#8581
We're supposed to have jupyter-offlinenotebook extension to be able to download notebooks after timeout.
It Currently does not work. After inactivity we end up with :
Despite :
jupyter-offlinenotebook in environment.ymljupyter labextension install jupyter-offlinenotebook in postBuildEdit : alternative timeout if i was in the "binder" folder [confirms that the "directory not bound" is the current directory]
Not supported in Jupyterlab 3.0 :
A declarative, efficient, and flexible JavaScript library for building user interfaces.
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
An Open Source Machine Learning Framework for Everyone
The Web framework for perfectionists with deadlines.
A PHP framework for web artisans
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
Some thing interesting about web. New door for the world.
A server is a program made to process requests and deliver data to clients.
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
Some thing interesting about visualization, use data art
Some thing interesting about game, make everyone happy.
We are working to build community through open source technology. NB: members must have two-factor auth.
Open source projects and samples from Microsoft.
Google ❤️ Open Source for everyone.
Alibaba Open Source for everyone
Data-Driven Documents codes.
China tencent open source team.