There are some non-tetrahedral stereochemistry annotations that are ignored in standardization. I wouldn't call these bugs, as they're somewhat obscure, and generally, have poor support in existing drawing/encoding software. However, I think they're important to note and worthwhile to investigate. I have included sd files for each case with its enantiomer in the tests folder.
Allene-Like Stereochemistry
JChem Smiles : Not supported
Daylight Smiles : Supported
InChi: Supported (via molfile)
Example: Mycomycin
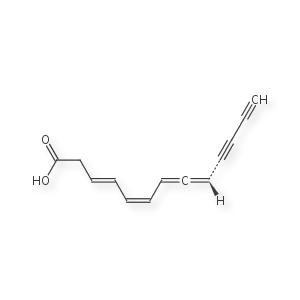
This is a special case, often described as tetrahedral stereo stretched out across two consecutive double bonds (allene). The defacto standard for drawing this configuration is to use dash/wedge for one side of the allene, and cis/trans like configuration on the other side. InChi respects this convention and will generate 2 different keys if I invert the dashes and wedges. Daylight smiles also allows this to be encoded (according to their website) but most tools I use either break or ignore their published rules.
Daylight smiles:
OC(C/C=C/C=C\C([H])=[C@]=C([H])C#CC#C)=O
Molfile:
Ketcher 12201302422D 1 1.00000 0.00000 0
17 16 0 0 0 999 V2000
-0.5000 -0.8660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.5000 0.8660 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.5000 -0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5000 -0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0000 -1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0000 -1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.5000 -0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
5.5000 -0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
6.5000 -0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
7.5000 0.8660 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.0000 1.7321 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
8.5000 2.5981 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0000 0.0000 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
7.0000 -1.7320 0.0000 H 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0
2 3 2 0 0 0
2 4 1 0 0 0
4 5 1 0 0 0
5 6 2 0 0 0
6 7 1 0 0 0
7 8 2 0 0 0
8 9 1 0 0 0
9 10 2 0 0 0
10 11 2 0 0 0
11 12 1 6 0 0
12 13 3 0 0 0
13 14 1 0 0 0
14 15 3 0 0 0
9 16 1 0 0 0
11 17 1 1 0 0
M END
Square-Planar Stereochemistry
JChem Smiles : Not supported
Daylight Smiles : Supported
InChi: Not supported
Example: Cisplatinin
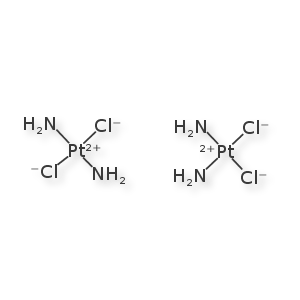
The left is cisplatinin, the right is transplatinin. They are distinct molecules that behave very differently in the clinic. And yet they are rarely treated as distinct by toolkits and registration systems. The square-planar stereochemistry is very straight-forward to draw. However, 1-D encoding and graph invariant annotations are undersupported for this class. This is one of the simpler extensions into inorganic chemistry that could be accomplished, and still, unfortunately, requires a bit of groundwork. Daylight's website claims to support this, but, again, I haven't found something to accept their encoding.
Daylight Smiles:
(N)[Pt@@SP1+2](N)([Cl-])[Cl-]
Molfile:
-OEChem-12201301332D
5 4 0 0 0 0 0 0 0999 V2000
0.0000 0.0000 0.0000 Pt 0 2 0 0 0 0 0 0 0 0 0 0
1.0308 -1.0897 0.0000 Cl 0 5 0 0 0 0 0 0 0 0 0 0
1.0897 1.0308 0.0000 Cl 0 5 0 0 0 0 0 0 0 0 0 0
-1.0308 1.0897 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
-1.0897 -1.0308 0.0000 N 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
1 3 1 0 0 0 0
1 4 1 0 0 0 0
1 5 1 0 0 0 0
M CHG 3 1 2 2 -1 3 -1
M END
Restricted Rotation Axial Stereochemistry
JChem Smiles : Not supported
Daylight Smiles : Not supported
InChi: Not supported
Example: R-GOSSYPOL
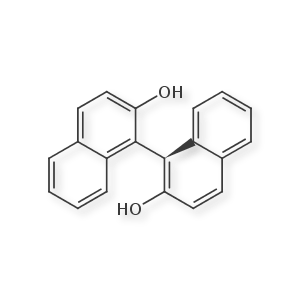
This happens in the special case where two phenyl rings are connected via a single bond, and both have sufficiently sized ortho substituents to restrict free rotation. Conceptually, this is similar to allene-like stereochemistry, in that the "stereo center" occurs across an axis rather than at a specific atom. However, I have found no 1D encoding of this form, and most molfile representations will try to overuse wedge bonds or non-standard "thicker" bonds to emphasize 3 dimensionality (much like with morphine). From my view, a single wedge/dash inside one of the aromatic rings in the molfile is ugly, but sufficient for annotation. If anyone is aware of accepted standards on drawing / encoding this, please let me know. I'd love to learn of a simple smiles extension that would encode this.
Molfile:
Ketcher 12201301552D 1 1.00000 0.00000 0
22 25 0 0 0 999 V2000
0.8660 1.5000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8660 1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7321 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7321 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5981 -0.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-2.5981 -1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-1.7321 -2.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8660 -1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
-0.8660 -0.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8660 -0.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.8660 -1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 -2.0000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.7321 -2.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5981 -1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5981 -0.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4641 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.4641 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.5981 1.5000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7321 1.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.7321 0.0000 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0
2 3 2 0 0 0
3 4 1 0 0 0
4 5 2 0 0 0
5 6 1 0 0 0
6 7 2 0 0 0
7 8 1 0 0 0
8 9 2 0 0 0
9 10 1 0 0 0
10 5 1 0 0 0
10 11 2 0 0 0
11 2 1 0 0 0
11 12 1 0 0 0
12 13 2 0 0 0
13 14 1 0 0 0
13 15 1 0 0 0
15 16 2 0 0 0
16 17 1 0 0 0
17 18 2 0 0 0
18 19 1 0 0 0
19 20 2 0 0 0
20 21 1 0 0 0
21 22 2 0 0 0
12 22 1 1 0 0
22 17 1 0 0 0
M END





















