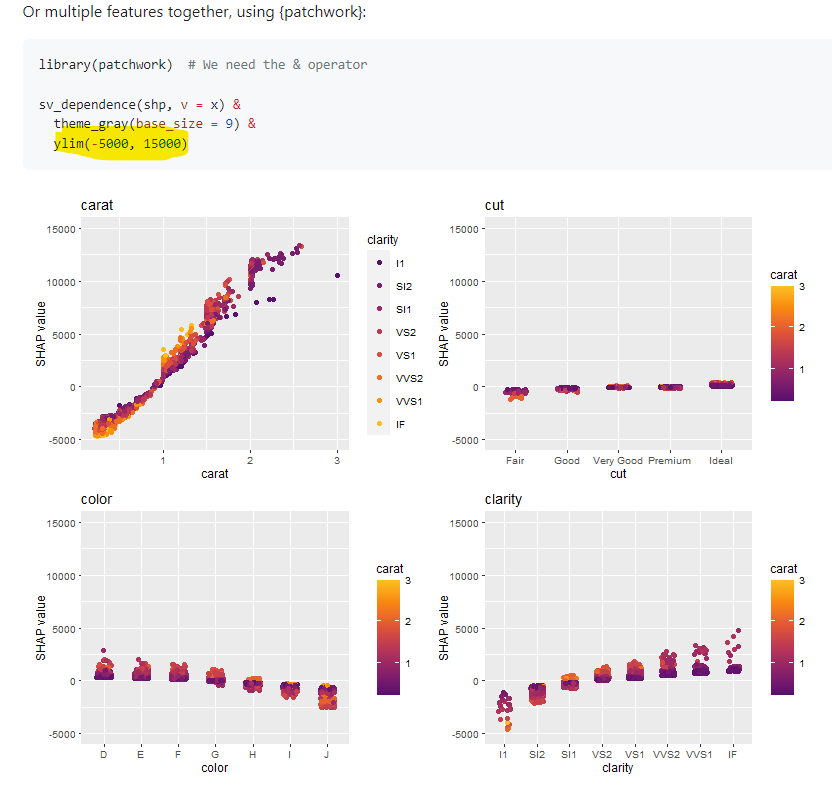
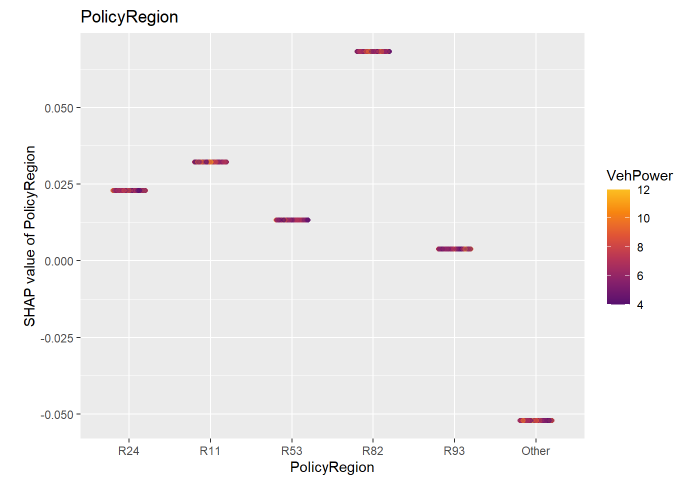
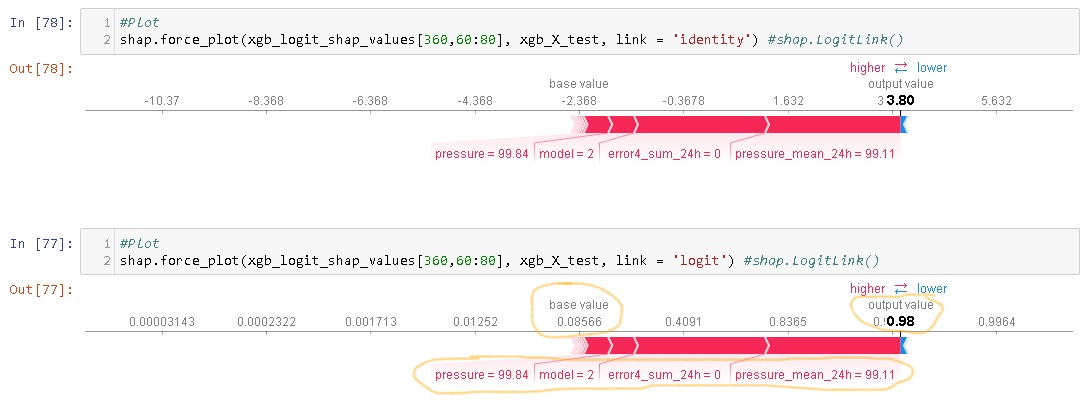
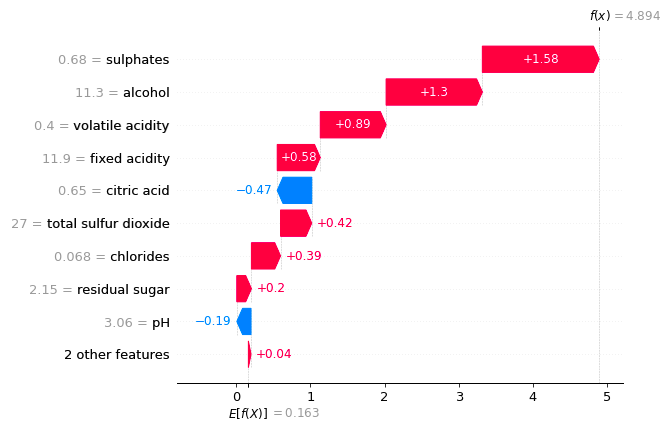
I think it would be useful to have a function that computes/visualises the relative importance of interaction effects.
Here's an example for an xgboost model where SHAP interaction values are available:
library(shapviz)
library(tidyverse)
library(xgboost)
set.seed(3653)
x <- c("carat", "cut", "color", "clarity")
dtrain <- xgb.DMatrix(data.matrix(diamonds[x]), label = diamonds$price)
fit <- xgb.train(params = list(learning_rate = 0.1), data = dtrain, nrounds = 65)
# Explanation data
dia_small <- diamonds[sample(nrow(diamonds), 2000), ]
# shapviz object with SHAP interaction values
shp_i <- shapviz(
fit, X_pred = data.matrix(dia_small[x]), X = dia_small, interactions = TRUE
)
# Interaction importance
shap_interactions <- apply(2 * abs(shp_i$S_inter), c(2, 3), mean)
shap_interactions[lower.tri(shap_interactions, diag = TRUE)] <- NA
as.data.frame.table(shap_interactions, responseName = "interaction_strength") %>%
filter(!is.na(interaction_strength)) %>%
arrange(desc(interaction_strength))
#> Var1 Var2 interaction_strength
#> 1 carat clarity 600.07087
#> 2 carat color 412.44253
#> 3 color clarity 188.35864
#> 4 carat cut 98.98317
#> 5 cut clarity 23.92846
#> 6 cut color 17.94669Created on 2023-10-24 with reprex v2.0.2
Ideally, this function would also work, based on some heuristics, for models that don't have SHAP interaction values available. I don't think using the heuristics in potential_interactions() (weighted squared correlations) willl work here as it doesn't take the amount of variation of the SHAP values in each bin into account, so the current interaction importance values are not comparable across features.
Maybe switching to the modelled part of the variation would work (and note that this also addresses #119): in each bin, fit a linear regression model and compute the mean of the absolute values of the fitted values minus the overall mean. I believe this boils down to the SHAP importance metric for a linear regression model with one feature. Doing so brings it on a scale that's comparable across bins and across features (differente vs in potential_interactions()).
Here's a code example to illustrate what I mean more clearly:
# shapviz object without interactions
shp <- shapviz(
fit, X_pred = data.matrix(dia_small[x]), X = dia_small, interactions = FALSE
)
# Replace correlation measure with modelled variation measure
# Swapping out the function `r_sq` with `mod_var`
potential_interactions_modelled <- function(obj, v) {
stopifnot(is.shapviz(obj))
S <- get_shap_values(obj)
S_inter <- get_shap_interactions(obj)
X <- get_feature_values(obj)
nms <- colnames(obj)
v_other <- setdiff(nms, v)
stopifnot(v %in% nms)
if (ncol(obj) <= 1L) {
return(NULL)
}
# Simple case: we have SHAP interaction values
if (!is.null(S_inter)) {
return(sort(2 * colMeans(abs(S_inter[, v, ]))[v_other], decreasing = TRUE))
}
# Complicated case: we need to rely on modelled variation based heuristic
mod_var <- function(s, x) {
sapply(x,
function(x) {
tryCatch({
mean(abs(stats::lm(s ~ x)$fitted - mean(s)))
}, error = function(e) {
return(NA)
})
})
}
n_bins <- ceiling(min(sqrt(nrow(X)), nrow(X) / 20))
v_bin <- shapviz:::.fast_bin(X[[v]], n_bins = n_bins)
s_bin <- split(S[, v], v_bin)
X_bin <- split(X[v_other], v_bin)
w <- do.call(rbind, lapply(X_bin, function(z) colSums(!is.na(z))))
modelled_variation <- do.call(rbind, mapply(mod_var, s_bin, X_bin, SIMPLIFY = FALSE))
sort(colSums(w * modelled_variation, na.rm = TRUE) / colSums(w), decreasing = TRUE)
}
# Current implementation with SHAP interaction values
potential_interactions(shp_i, v = "cut")
#> carat clarity color
#> 98.98315 23.92846 17.94669
# Current implementation based on heuristics
potential_interactions(shp, v = "cut")
#> carat clarity color
#> 0.49739669 0.07223855 0.04243011
# Suggested implementation based on heuristics
potential_interactions_modelled(shp, v = "cut")
#> carat clarity color
#> 35.23818 14.73922 10.66934
# Current implementation with SHAP interaction values
potential_interactions(shp_i, v = "carat")
#> clarity color cut
#> 600.07087 412.44253 98.98317
# Current implementation based on heuristics
potential_interactions(shp, v = "carat")
#> clarity color cut
#> 0.5301601 0.1545190 0.1121987
# Suggested implementation based on heuristics
potential_interactions_modelled(shp, v = "carat")
#> clarity color cut
#> 248.3854 177.3795 132.6098
# Function to create table with ranked interaction variables
table_potential_interactions <- function(predictor) {
pi <- potential_interactions_modelled(shp, predictor)
tibble(var1 = predictor, var2 = names(pi), interaction_strength = pi)
}
# Interaction importance
map(x, table_potential_interactions) %>%
bind_rows() %>%
arrange(desc(interaction_strength))
#> # A tibble: 12 × 3
#> var1 var2 interaction_strength
#> <chr> <chr> <dbl>
#> 1 carat clarity 248.
#> 2 clarity carat 186.
#> 3 carat color 177.
#> 4 carat cut 133.
#> 5 color carat 128.
#> 6 clarity color 89.3
#> 7 color clarity 55.6
#> 8 clarity cut 48.7
#> 9 cut carat 35.2
#> 10 color cut 25.0
#> 11 cut clarity 14.7
#> 12 cut color 10.7Note that this analysis is not symmetric, but I don't think that's an issue as the table above is informative: it suggests you to split out var1 effects by var2 and hence look at PD plots or SHAP dependence plots for var1 by different segments of var2.