Comments (4)
Inferring taxonomy for metagenomes
Note
The names in the output of the following two commands is not the same as on the tutorial page Ribosomal_S6_2915 (Tutorial) Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655 (my output).
command
anvi-estimate-scg-taxonomy -c CONTIGS.db \
--metagenome-mode```
output
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| | percent_identity | taxonomy |
+==========================================================+====================+=================================================================================================================================+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / Staphylococcus epidermidis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_12163 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / Staphylococcus hominis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_15200 | 98.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Lactobacillaceae / Leuconostoc / Leuconostoc citreum |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_25880 | 98.9 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Finegoldia / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_2915 | 97.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Enterococcaceae / Enterococcus / Enterococcus faecalis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_29818 | 97.9 | Bacteria / Firmicutes / Bacilli / Lactobacillales / Streptococcaceae / Streptococcus / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_30904 | 98.1 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Anaerococcus / Anaerococcus nagyae |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_4484 | 98.9 | Bacteria / Firmicutes / Clostridia / Tissierellales / Peptoniphilaceae / Peptoniphilus / Peptoniphilus lacydonensis |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_6421 | 100 | Bacteria / Actinobacteriota / Actinomycetia / Propionibacteriales / Propionibacteriaceae / Cutibacterium / Cutibacterium avidum |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_7660 | 98.9 | Bacteria / Firmicutes / Bacilli / Staphylococcales / Staphylococcaceae / Staphylococcus / |
+----------------------------------------------------------+--------------------+---------------------------------------------------------------------------------------------------------------------------------+
command
anvi-estimate-scg-taxonomy -c CONTIGS.db \
> -p PROFILE.db \
> --metagenome-mode \
> --compute-scg-coverages
output
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| | percent_identity | taxonomy | DAY_15A | DAY_15B | DAY_16 | DAY_17A | DAY_17B | ... 6 more |
+==========================================================+====================+================================+===========+===========+==========+===========+===========+==============+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_2915 | 97.9 | (s) Enterococcus faecalis | 372.512 | 699.853 | 663.241 | 186.34 | 1149.6 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_11655 | 98.9 | (s) Staphylococcus epidermidis | 112.694 | 172.478 | 147.304 | 23.3901 | 140.769 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_4484 | 98.9 | (s) Peptoniphilus lacydonensis | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_7660 | 98.9 | (g) Staphylococcus | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_6421 | 100 | (s) Cutibacterium avidum | 17.8935 | 5.87368 | 4.79909 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_12163 | 98.9 | (s) Staphylococcus hominis | 2.39322 | 22.5447 | 13.2806 | 0 | 9.94853 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_15200 | 98.9 | (s) Leuconostoc citreum | 0 | 0 | 1.86532 | 0 | 1.6936 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_29818 | 97.9 | (g) Streptococcus | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_25880 | 98.9 | (g) Finegoldia | 0 | 0 | 0 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
| Infant_Gut_Contigs_from_Sharon_et_al__Ribosomal_S6_30904 | 98.1 | (s) Anaerococcus nagyae | 0.528958 | 0 | 0.14157 | 0 | 0 | ... 6 more |
+----------------------------------------------------------+--------------------+--------------------------------+-----------+-----------+----------+-----------+-----------+--------------+
from merenlab.org.
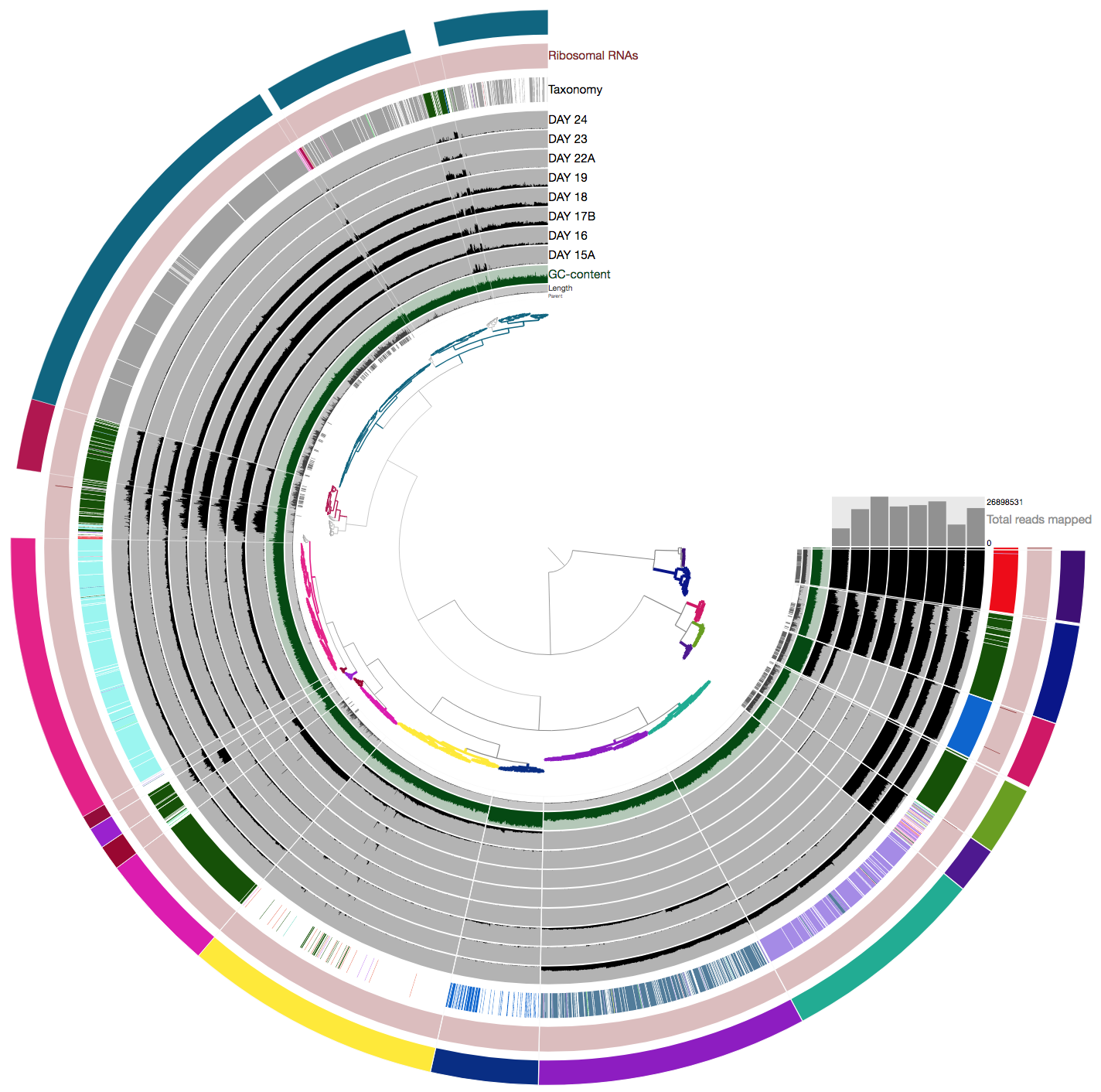
Manual identification of genomes in the Infant Gut Dataset
Note
For the manual identification of bins it would be helpful for the screenshot to include the information in the Bins tab (Len, Comp., Red.)
Image from tutorial
Image from my interface (please disregard that the bins I assigned are slightly different from the tutorial)
from merenlab.org.
Chapter II: Automatic Binning
Manually curating automatic binning outputs
Note
Minor note, but I got a slightly different redundancy value than you report in the text (maxbin_007 has a redundancy value of 22% in the tutorial, 18.31% for me)
Text in tutorial
It is clear that some bins are not as well-resolved as others. For instance, bins maxbin_007 and maxbin_008 have redundancy estimates of 22% and 91%, respectively, which suggests each of them describe multiple distinct populations. Well, clearly we would have preferred those bins to behave.
command
anvi-estimate-genome-completeness -p PROFILE.db \
-c CONTIGS.db \
-C MAXBIN
my output
Bins in collection "MAXBIN"
===============================================
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| bin name | domain | confidence | % completion | % redundancy | num_splits | total length |
+============+==========+==============+================+================+==============+================+
| maxbin_001 | BACTERIA | 1 | 100 | 4.23 | 148 | 2969341 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_002 | BACTERIA | 1 | 94.37 | 0 | 88 | 1801068 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_003 | BACTERIA | 0.7 | 81.69 | 2.82 | 144 | 2764617 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_004 | BACTERIA | 1 | 97.18 | 1.41 | 188 | 2571878 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_005 | BACTERIA | 1 | 98.59 | 0 | 151 | 2555414 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_006 | BACTERIA | 0.7 | 78.87 | 5.63 | 305 | 2901149 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_007 | BACTERIA | 0.4 | 54.93 | 18.31 | 724 | 3230717 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_008 | BACTERIA | 0.8 | 87.32 | 90.14 | 1657 | 3056818 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
| maxbin_009 | EUKARYA | 0.8 | 83.13 | 7.23 | 1379 | 13915165 |
+------------+----------+--------------+----------------+----------------+--------------+----------------+
from merenlab.org.
Thanks for the heads up on these issues @raissameyer :) I started a branch (IGT_fixes_from_Raissa) and fixed the table outputs. I didn't want to update the binning screenshot myself since it has nice colors, so I leave that one to @metehaansever :)
from merenlab.org.
Related Issues (6)
- minor blog issues HOT 1
- ToC cursor HOT 1
- Are hyperlinks hard to see? HOT 6
- Ncbi-genome-download HOT 2
- error in gff_parser.py HOT 1
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.

from merenlab.org.