By Johann Brehmer, Felix Kling, Irina Espejo, Sinclert Pérez, and Kyle Cranmer
Particle physics processes are usually modeled with complex Monte-Carlo simulations of the hard process, parton shower, and detector interactions. These simulators typically do not admit a tractable likelihood function: given a (potentially high-dimensional) set of observables, it is usually not possible to calculate the probability of these observables for some model parameters. Particle physicists usually tackle this problem of "likelihood-free inference" by hand-picking a few "good" observables or summary statistics and filling histograms of them. But this conventional approach discards the information in all other observables and often does not scale well to high-dimensional problems.
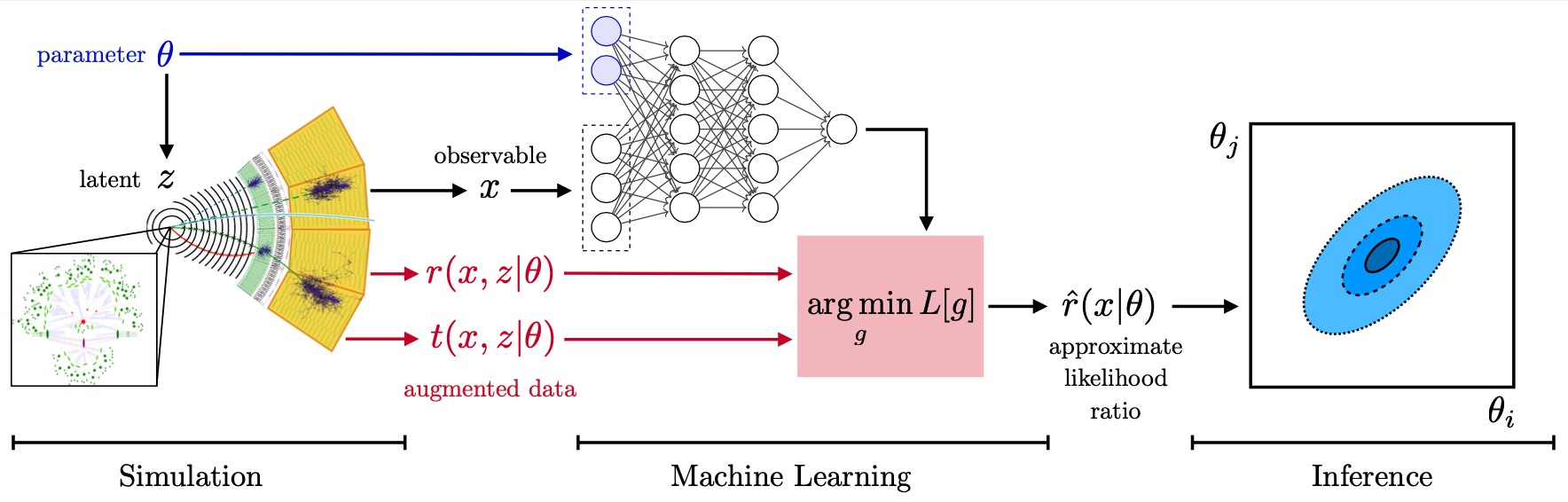
In the three publications "Constraining Effective Field Theories with Machine Learning", "A Guide to Constraining Effective Field Theories with Machine Learning", and "Mining gold from implicit models to improve likelihood-free inference", a new approach has been developed. In a nutshell, additional information is extracted from the simulations that is closely related to the matrix elements that determine the hard process. This "augmented data" can be used to train neural networks to efficiently approximate arbitrary likelihood ratios. We playfully call this process "mining gold" from the simulator, since this information may be hard to get, but turns out to be very valuable for inference.
But the gold does not have to be hard to mine: MadMiner automates these modern multivariate inference strategies. It wraps around the simulators MadGraph and Pythia, with different options for the detector simulation. It streamlines all steps in the analysis chain from the simulation to the extraction of the augmented data, their processing, the training and evaluation of the neural networks, and the statistical analysis are implemented.
Our main publication MadMiner: Machine-learning-based inference for particle physics provides an overview over this package. We recommend reading it first before jumping into the code.
Please have a look at our installation instructions.
In the examples folder in this repository, we provide two tutorials. The first is called Toy simulator, and it is based on a toy problem rather than a full particle-physics simulation. It demonstrates inference with MadMiner without spending much time on the more technical steps of running the simulation. The second, called Particle physics, shows all steps of a particle-physics analysis with MadMiner.
These examples are the basis of the online tutorial built on Jupyter Books. It also walks through how to run MadMiner using Docker so that you do not have to install Fortran, MadGraph, Pythia, Delphes, etc. You can even run it with no install using Binder.
The madminer API is documented on Read the Docs.
If you have any questions, please chat to us in our Gitter community.
If you use MadMiner, please cite our main publication,
@article{Brehmer:2019xox,
author = "Brehmer, Johann and Kling, Felix and Espejo, Irina and Cranmer, Kyle",
title = "{MadMiner: Machine learning-based inference for particle physics}",
journal = "Comput. Softw. Big Sci.",
volume = "4",
year = "2020",
number = "1",
pages = "3",
doi = "10.1007/s41781-020-0035-2",
eprint = "1907.10621",
archivePrefix = "arXiv",
primaryClass = "hep-ph",
SLACcitation = "%%CITATION = ARXIV:1907.10621;%%"
}
The code itself can be cited as
@misc{MadMiner_code,
author = "Brehmer, Johann and Kling, Felix and Espejo, Irina and Perez, Sinclert and Cranmer, Kyle",
title = "{MadMiner}",
doi = "10.5281/zenodo.1489147",
url = {https://github.com/madminer-tool/madminer}
}
The main references for the implemented inference techniques are the following:
- CARL: 1506.02169.
- MAF: 1705.07057.
- CASCAL, RASCAL, ROLR, SALLY, SALLINO, SCANDAL:
- ALICE, ALICES: 1808.00973.
We are immensely grateful to all contributors and bug reporters! In particular, we would like to thank Zubair Bhatti, Philipp Englert, Lukas Heinrich, Alexander Held, Samuel Homiller and Duccio Pappadopulo.
The SCANDAL inference method is based on Masked Autoregressive Flows, where our implementation is a PyTorch port of the original code by George Papamakarios, available at this repository.