One important advantage in plant&animal genetic studies is the availability of experimental populations. A Recombinant Inbred Line (RIL) population can be maintained and used over and over again to map all kinds of different traits. Now, RIL population is not confined to bi-parental RIL population. It can be nested association mapping (NAM) population or multiparent advanced generation intercross (MAGIC) population of arbitrary number of founders. I present a method BCFR for constructing bin-map for RILs of arbitrary number of founders. BCFR use identity by state between RILs and their founders in sliding windows, and a pedigree relationship can be optionally used to refine the bin-map, that is, individuals derived from single F1 hybrid can only have two types of alleles.
-
Python (>=3.5.5)
-
Click (>=7.0)
-
Numpy (>=1.13.1)
-
Pandas (>=0.21.0)
-
Matplotlib (>=1.5.0)
Unzip the bcfr code and install
unzip bcfr-master.zip
cd bcfr-master
python setup.py install
List all the options and commands:
bcfr --help
Convert the VCF file to bcfr format:
bcfr converter --v my_genotypes.vcf --d output_dir --n number_of_founder
Construct bin map without pedigree:
bcfr birds --n number_of_founder --d output_dir
Construct bin map with pedigree:
bcfr birds --n number_of_founder --d output_dir --p pedigree.txt
Plot individual haplotype map:
bcfr plot-hap --r ril_individual_name --d output_dir
Note: the output_dir should be the same directory in all these steps.
pedigree template was offered as foder.txt
bimDic.pkl is the pickled bin map object created by birds command which can be loaded in python:
with open('bimDic.pkl','rb') as f:
bin_size,bin_map=pickle.load(f)
bin_size and bin_map are dictionary with chromosome names as key. bin_size stores the start position and end position of each bin; bin_map stores the genotypes.
The individual-wise haplotype map data was hpfDic.pkl.
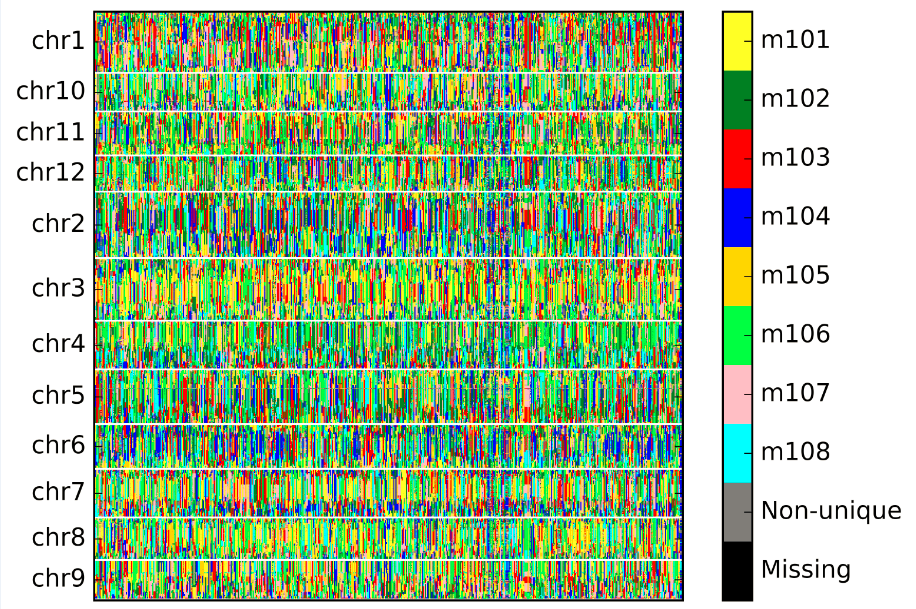
The whole genome bin map chart was output as whole_genome_bin_map.jpg:
The plot-hap command output individual haplotype map chart haplotype_ril_individual_name.jpg:
Zhongmin Han - National Key Laboratory of Crop Genetic Improvement, Huazhong Agriculture University
Bin-based genome-wide association analyses improve power and resolution in QTL mapping and identify favorable alleles from multiple parents in a 4-way MAGIC rice population. Theoretical and Applied Genetics by Zhongmin Han, Gang Hu, Hua Liu, Famao Liang, Lin Yang, Hu Zhao, Qinghua Zhang, Zhixin Li, Qifa Zhang, Yongzhong Xing
- Lots of memory and fast disk for large projects