Dear Max Gordon,
I regularly use your package, which I find very practical and sometimes more flexible than others. Thank you for its development !
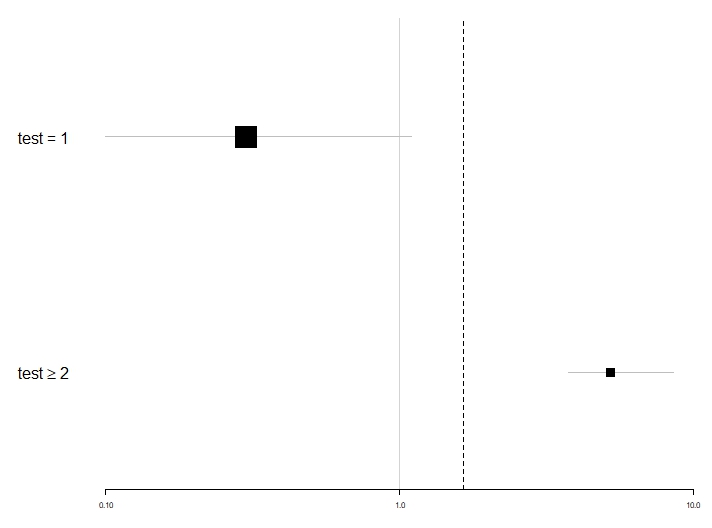
Recently, I've been experiencing difficulties to produce a foresplot and I don't know what could be the reason for this. Could you tell me if this is due to a misuse of your package.
Below are the parameters of my session. I also attach a capture of the resulting graph, the script and the data extract used.

db <- structure(list(varname = c("Restriction de participation", " Hommes",
" Femmes", "Temps d'activité réduit", " Hommes", " Femmes",
"Faible niveau d'éducation", " Hommes", " Femmes", "Sans emploi",
" Hommes", " Femmes", "Ménage pauvre", " Hommes", " Femmes",
"Confiage dans l'enfance", " Hommes", " Femmes", "Faible soutien social",
" Hommes", " Femmes", " "), coef = c("", "1.13", "0.96",
"", "3.22", "0.85", "", "1.72", "0.99", "", "-0.84", "-0.25",
"", "0.65", "0.54", "", "-0.02", "0.19", "", "0.75", "0.76",
""), ci.low = c("", "0.728", "0.575", "", "2.31", "0.434", "",
"1.296", "0.601", "", "-1.401", "-0.78", "", "0.195", "0.085",
"", "-0.446", "-0.206", "", "0.298", "0.355", ""), ci.high = c("",
"1.535", "1.36", "", "4.432", "1.271", "", "2.164", "1.392",
"", "-0.293", "0.272", "", "1.116", "1.001", "", "0.399", "0.582",
"", "1.213", "1.18", ""), pval = c("", "<0.001", "<0.001", "",
"<0.001", "<0.001", "", "<0.001", "<0.001", "", "0.003", "0.348",
"", "0.006", "0.021", "", "0.913", "0.352", "", "0.001", "<0.001",
""), nlab.ph = c("", "123 (59)", "134 (63)", "", "68 (33)", "93 (44)",
"", "126 (61)", "120 (57)", "", "163 (78)", "171 (81)", "", "63 (30)",
"60 (28)", "", "61 (29)", "85 (40)", "", "67 (32)", "91 (43)",
""), nlab.pt = c("", "67 (32)", "84 (40)", "", "4 (2)", "55 (26)",
"", "45 (22)", "69 (33)", "", "184 (88)", "178 (84)", "", "39 (19)",
"40 (19)", "", "62 (30)", "76 (36)", "", "39 (19)", "55 (26)",
""), status.lab = c("Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)",
"Personnes handicapées (PH) vs Personnes non handicapées (PnH)"
), coef.lab = c("", "1.13[0.728;1.535]", "0.96[0.575;1.36]",
"", "3.22[2.31;4.432]", "0.85[0.434;1.271]", "", "1.72[1.296;2.164]",
"0.99[0.601;1.392]", "", "-0.84[-1.401;-0.293]", "-0.25[-0.78;0.272]",
"", "0.65[0.195;1.116]", "0.54[0.085;1.001]", "", "-0.02[-0.446;0.399]",
"0.19[-0.206;0.582]", "", "0.75[0.298;1.213]", "0.76[0.355;1.18]",
"")), row.names = c(NA, -22L), class = c("tbl_df", "tbl", "data.frame"
))
table.text <- cbind(
c("Caracteristique",db$varname),
c("PH (%)",db$nlab.ph),
c("PnH (%)",db$nlab.pt),
c("Coef",db$coef),
c("p-value",db$pval)
)
forestplot(labeltext=table.text,
title="PH vs PnH",
fn.ci_norm = c(fpDrawNormalCI,#Entete
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Restriction de participation
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Temps d'activité réduit
fpDrawNormalCI,fpDrawNormalCI,fpDrawCircleCI, #Faible niveau d'éducation
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Sans emploi
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Ménage pauvre
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Confiage dans l'enfance
fpDrawNormalCI,fpDrawCircleCI,fpDrawNormalCI, #Faible soutien social
fpDrawNormalCI), #Pieds de page
xlab="Coefficient adjusté sur le groupe d'âge (Coef)",
graph.pos = ncol(table.text),
mean=c(NA,db$coef),
lower=c(NA,db$ci.low),
upper=c(NA,db$ci.high),
is.summary = c(TRUE,rep(FALSE,nrow(table.text)-2),TRUE),
hrzl_lines=list("2"=gpar(lty=1,lwd=2),
"23"=gpar(lty=2,lwd=1,columns=c(1:(ncol(table.text)-1),ncol(table.text)+1))),
zero=0,line.margin = .1,cex=0.9,lwd.ci=2,boxsize = .25,
ci.vertices = TRUE, colgap=unit(3,"mm"),
clip=c(-2,5),xticks = c(-2,-1.5,-1,-0.5,0,0.5,1,1.5,2,2.5,3,3.5,4,4.5,5),
grid=gpar(lty=3,lwd=1,col="gray75"),
txt_gp=fpTxtGp(label = gpar(cex=0.75),ticks=gpar(cex=0.5),xlab = gpar(cex=0.6,fontface="bold")),
summary=list("1"=list(gpar(cex=0.7,fontface="bold"),gpar(cex=0.7,fontface="bold"),
gpar(cex=0.7,fontface="bold"),gpar(cex=0.7,fontface="bold"),
gpar(cex=0.65,fontface="bold")),
"23"=list(gpar(cex=0.5,fontface="italic"),gpar(cex=0.5,fontface="italic"),
gpar(cex=0.5,fontface="italic"),gpar(cex=0.5,fontface="italic"),
gpar(cex=0.5,fontface="italic"))),
col=fpColors(box="black", lines="black", zero = "gray25",summary="black")
)
R version 3.5.3 (2019-03-11)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.15.3
Matrix products: default
BLAS: /System/Library/Frameworks/Accelerate.framework/Versions/A/Frameworks/vecLib.framework/Versions/A/libBLAS.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] grid stats graphics grDevices utils datasets methods base
other attached packages:
[1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.4 purrr_0.3.3 readr_1.3.1 tidyr_1.0.2
[7] tibble_2.1.3 tidyverse_1.3.0 ggplot2_3.2.1 ggpubr_0.2.4 finalfit_0.9.7 forestplot_1.9
[13] FactoMineR_2.0 checkmate_2.0.0 texreg_1.36.23 magrittr_1.5
loaded via a namespace (and not attached):
[1] httr_1.4.1 jsonlite_1.6.1 splines_3.5.3 modelr_0.1.5 rmeta_3.0
[6] Formula_1.2-3 assertthat_0.2.1 latticeExtra_0.6-28 cellranger_1.1.0 ggrepel_0.8.1
[11] pillar_1.4.3 backports_1.1.5 lattice_0.20-38 glue_1.3.1 digest_0.6.25
[16] RColorBrewer_1.1-2 ggsignif_0.6.0 rvest_0.3.5 minqa_1.2.4 colorspace_1.4-1
[21] htmltools_0.4.0 Matrix_1.2-18 pkgconfig_2.0.3 broom_0.5.4 haven_2.2.0
[26] scales_1.1.0 dummies_1.5.6 lme4_1.1-21 htmlTable_1.13.3 generics_0.0.2
[31] withr_2.1.2 pan_1.6 nnet_7.3-12 lazyeval_0.2.2 cli_2.0.1
[36] readxl_1.3.1 survival_3.1-8 crayon_1.3.4 mitml_0.3-7 mice_3.7.0
[41] fansi_0.4.1 fs_1.3.1 nlme_3.1-143 MASS_7.3-51.4 xml2_1.2.2
[46] foreign_0.8-72 tools_3.5.3 data.table_1.12.8 hms_0.5.3 lifecycle_0.1.0
[51] reprex_0.3.0 munsell_0.5.0 cluster_2.1.0 packrat_0.5.0 flashClust_1.01-2
[56] compiler_3.5.3 rlang_0.4.4 nloptr_1.2.1 rstudioapi_0.11 htmlwidgets_1.5.1
[61] leaps_3.1 base64enc_0.1-3 boot_1.3-23 gtable_0.3.0 DBI_1.0.0
[66] R6_2.4.1 lubridate_1.7.4 gridExtra_2.3 knitr_1.26 Hmisc_4.3-0
[71] jomo_2.6-10 stringi_1.4.6 parallel_3.5.3 Rcpp_1.0.3 vctrs_0.2.3
[76] rpart_4.1-15 acepack_1.4.1 scatterplot3d_0.3-41 dbplyr_1.4.2 tidyselect_1.0.0
[81] xfun_0.11