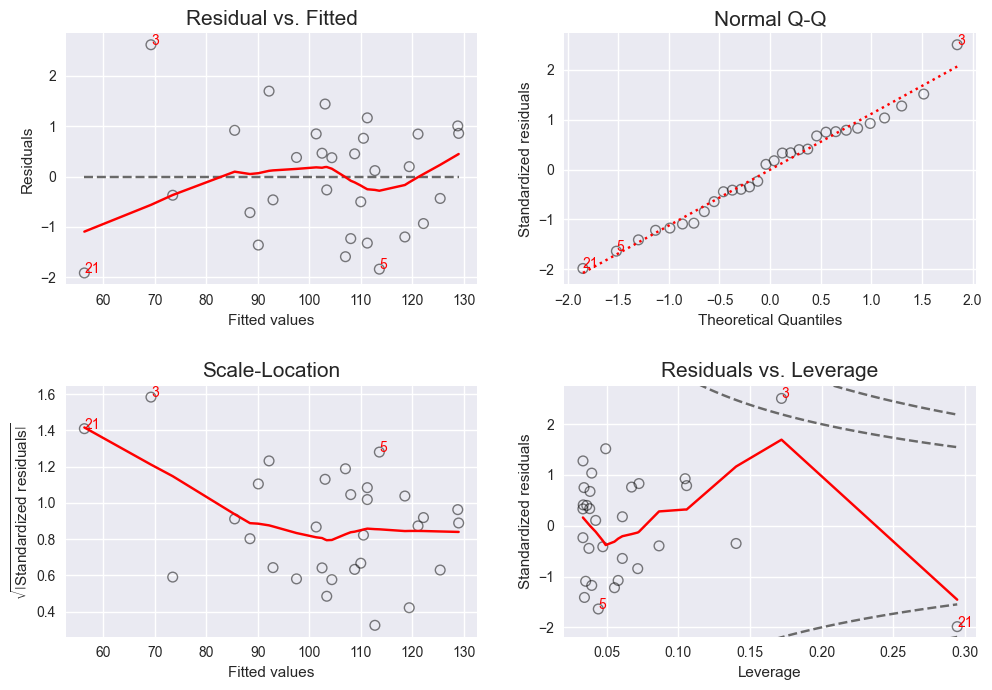
Python Library providing Diagnostic Plots for Linear Regression Models. (Like plot.lm in R.)
I built this, because I missed the diagnostics plots of R for a university project. There are some substitutions in Python for individual charts, but they are spread over different libraries and sometimes don't show the exact same. My implementation tries to copycat the R-plots, but I didn't reimplement the R-code: The charts are just based on available documentation.
pip install lmdiag
lmdiag generates plots for fitted linear regression models from
statsmodels,
linearmodels and
scikit-learn.
You can find some usage examples in this jupyter notebook.
import numpy as np
import statsmodels.api as sm
import lmdiag
# Fit model with random sample data
np.random.seed(20)
X = np.random.normal(size=30, loc=20, scale=3)
y = 5 + 5 * X + np.random.normal(size=30)
X = sm.add_constant(predictor) # intercept required by statsmodels
lm = sm.OLS(y, X).fit()
# Plot lmdiag facet chart
lmdiag.style.use(style="black_and_red") # Mimic R's plot.lm style
fig = lmdiag.plot(lm)
fig.show()-
Draw matrix of all plots:
lmdiag.plot(lm) -
Draw individual plots:
lmdiag.resid_fit(lm)lmdiag.q_q(lm)lmdiag.scale_loc(lm)lmdiag.resid_lev(lm) -
Print description to aid plot interpretation:
lmdiag.help()(for all plots)lmdiag.help('<method name>')(for individual plot)
Plotting models fitted on large datasets might be slow. There are some things you can try to speed it up:
The red smoothing lines are calculated using the "Locally Weighted Scatterplot
Smoothing" algorithm, which can be quite expensive. Try a lower value for lowess_it
and a higher value for lowess_delta to gain speed at the cost of accuracy:
lmdiag.plot(lm, lowess_it=1, lowess_delta=0.02)
# Defaults are: lowess_it=2, lowess_delta=0.005(For details about those parameters, see statsmodels docs.)
Try a different
matplotlib backend.
Especially static backends like AGG or Cairo should be faster, e.g.:
import matplotlib
matplotlib.use('agg')python -m venv .venv
source .venv/bin/activate
pip install -e '.[dev]'
pre-commit install