NOTE: dicompyler has been archived and is no longer supported. The developers of dicompyler and DVH Analytics have teamed up to form ONCurate.
dicompyler is an extensible open source radiation therapy research platform based on the DICOM standard. It also functions as a cross-platform DICOM RT viewer.dicompyler is written in Python and is built on a number of technologies including: pydicom, wxPython, Pillow, and matplotlib and runs on Windows, Mac OS X and Linux.
dicompyler is released under a BSD license.
Take a tour of dicompyler by checking out some screenshots or download a copy today.
Downloads are available through Google Drive:
Version 0.4.2: Windows | Mac | Source | Test Data - Released July 15th, 2014 - Release Notes
- Import CT/MR/PET Images, DICOM RT structure set, RT dose and RT plan files
- Extensible plugin system with included plugins:
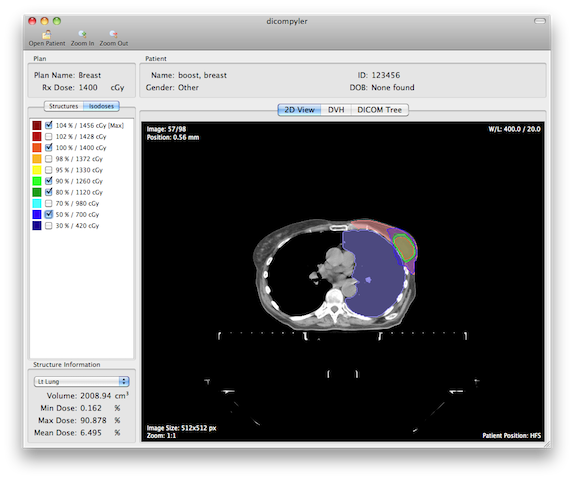
- 2D image viewer with dose and structure overlay
- Dose volume histogram viewer with the ability to analyze DVH parameters
- DICOM data tree viewer
- Patient anonymizer
- 3rd-party plugins can be found at https://github.com/dicompyler/dicompyler-plugins
- Custom plugins can be written by following the Plugin development guide
For upcoming features, see the project roadmap.
- Windows XP/Vista/7/8/10
- Mac OS X 10.5 - 10.13 (Intel) - Must bypass Gatekeeper
- Linux - via a package from PyPI or a Debian package (courtesy of debian-med)
If you are interested in building from source, please check out the build instructions.
- How to run dicompyler:
-
If you have downloaded dicompyler as an application for Windows or Mac, please follow the normal process for running any other application on your system.
-
If you are running from a Python package, a script called "dicompyler" will now be present on your path, which you can run from your command line or terminal.
-
If you are running from a source checkout, there is a script in the main folder called "dicompyler_app.py" which can be executed via your Python interpreter.
-
dicompyler will read properly formatted DICOM and DICOM-RT files. To get started, run dicompyler and click "Open Patient" to bring up a dialog box that will show the DICOM files in the last selected directory. You may click "Browse..." to navigate to other folders that contain DICOM data.
In the current version of dicompyler, you can import any DICOM CT, PET, or MRI image series, DICOM RT structure set, RT dose and RT plan files. dicompyler will automatically highlight the most dependent item for the patient. All related items (up the tree) will be automatically imported as well.
Alternatively, you can selectively import data. For example, If you only want to import CT images and an RT structure set just highlight the RT structure set. If you are importing an RT dose file and the corresponding plan does not contain a prescription dose, enter one in the box first. To import the data, click "Select" and dicompyler will process the information.
Once the DICOM data has been loaded, the main window will show the patient and plan information. Additionally it will show a list of structures and isodoses that are associated with the plan.
- As a starting point, please read the FAQ as it answers the most commonly asked questions about dicompyler.
- If you are unable to find the answer in the FAQ or in the wiki, dicompyler has a discussion forum hosted on Google Groups.
- If you need to cite dicompyler as a reference in your publication, please use the following citation:
- A Panchal and R Keyes. "SU-GG-T-260: dicompyler: An Open Source Radiation Therapy Research Platform with a Plugin Architecture" Med. Phys. 37, 3245, 2010
- The reference in Medical Physics can be accessed via http://dx.doi.org/10.1118/1.3468652