📚Documentation | 🛠️ Installation | 🌎 Home Page | 🐿🐴🐁🐘🐆 Model Zoo | 🚨 News | 🪲 Reporting Issues
🫶 Getting Assistance | ∞ DeepLabCut Online Course | 📝 Publications | 👩🏾💻👨💻 DeepLabCut AI Residency
DeepLabCut™️ is a toolbox for state-of-the-art markerless pose estimation of animals performing various behaviors. As long as you can see (label) what you want to track, you can use this toolbox, as it is animal and object agnostic. Read a short development and application summary below.
Please click the link above for all the information you need to get started! Please note that currently we support only Python 3.10+ (see conda files for guidance).
Developers Stable Release:
- Very quick start: You need to have TensorFlow installed (up to v2.10 supported across platforms)
pip install "deeplabcut[gui,tf]"that includes all functions plus GUIs, orpip install deeplabcut[tf](headless version with PyTorch and TensorFlow).
Developers Alpha Release:
- We also have an alpha release of PyTorch DeepLabCut available! Please see here for instructions and information.
We recommend using our conda file, see here or the new deeplabcut-docker package.
Our docs walk you through using DeepLabCut, and key API points. For an overview of the toolbox and workflow for project management, see our step-by-step at Nature Protocols paper.
For a deeper understanding and more resources for you to get started with Python and DeepLabCut, please check out our free online course! http://DLCcourse.deeplabcut.org
🐭 pose tracking of single animals demo
🐭🐭🐭 pose tracking of multiple animals demo
- See more demos here. We provide data and several Jupyter Notebooks: one that walks you through a demo dataset to test your installation, and another Notebook to run DeepLabCut from the beginning on your own data. We also show you how to use the code in Docker, and on Google Colab.
In 2018, we demonstrated the capabilities for trail tracking, reaching in mice and various Drosophila behaviors during egg-laying (see Mathis et al. for details). There is, however, nothing specific that makes the toolbox only applicable to these tasks and/or species. The toolbox has already been successfully applied (by us and others) to rats, humans, various fish species, bacteria, leeches, various robots, cheetahs, mouse whiskers and race horses. DeepLabCut utilized the feature detectors (ResNets + readout layers) of one of the state-of-the-art algorithms for human pose estimation by Insafutdinov et al., called DeeperCut, which inspired the name for our toolbox (see references below). Since this time, the package has changed substantially. The code has been re-tooled and re-factored since 2.1+: We have added faster and higher performance variants with MobileNetV2s, EfficientNets, and our own DLCRNet backbones (see Pretraining boosts out-of-domain robustness for pose estimation and Lauer et al 2022). Additionally, we have improved the inference speed and provided both additional and novel augmentation methods, added real-time, and multi-animal support. In v3.0+ we have changed the backend to support PyTorch. This brings not only an easier installation process for users, but performance gains, developer flexibility, and a lot of new tools! Importantly, the high-level API stays the same, so it will be a seamless transition for users 💜! We currently provide state-of-the-art performance for animal pose estimation and the labs (M. Mathis Lab and A. Mathis Group) have both top journal and computer vision conference papers.
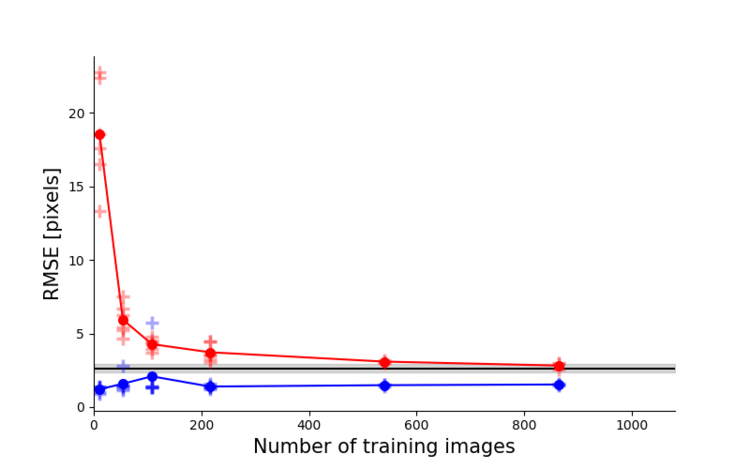
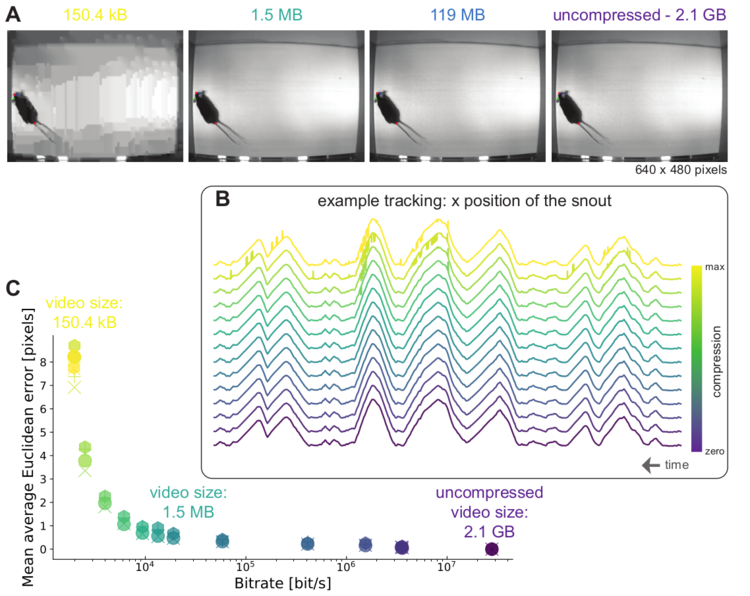
Left: Due to transfer learning it requires little training data for multiple, challenging behaviors (see Mathis et al. 2018 for details). Mid Left: The feature detectors are robust to video compression (see Mathis/Warren for details). Mid Right: It allows 3D pose estimation with a single network and camera (see Mathis/Warren). Right: It allows 3D pose estimation with a single network trained on data from multiple cameras together with standard triangulation methods (see Nath* and Mathis* et al. 2019).
DeepLabCut is embedding in a larger open-source eco-system, providing behavioral tracking for neuroscience, ecology, medical, and technical applications. Moreover, many new tools are being actively developed. See DLC-Utils for some helper code.
DLC code was originally developed by Alexander Mathis & Mackenzie Mathis, and was extended in 2.0 with the core dev team consisting of Tanmay Nath (2.0-2.1), and currently (2.1+) with Jessy Lauer and (2.3+) Niels Poulsen. DeepLabCut is an open-source tool and has benefited from suggestions and edits by many individuals including Mert Yuksekgonul, Tom Biasi, Richard Warren, Ronny Eichler, Hao Wu, Federico Claudi, Gary Kane and Jonny Saunders as well as the 100+ contributors. Please see AUTHORS for more details!
This is an actively developed package and we welcome community development and involvement.
| 🚉 Platform | 🎯 Goal | ⏱️ Estimated Response Time | 📢 Support Squad |
|---|---|---|---|
 🐭Tag: DeepLabCut |
To ask help and support questions👋 | Promptly🔥 | DLC Team and The DLC Community |
| GitHub DeepLabCut/Issues | To report bugs and code issues🐛 (we encourage you to search issues first) | 2-3 days | DLC Team |
| To discuss with other users, share ideas and collaborate💡 | 2 days | The DLC Community | |
| GitHub DeepLabCut/Contributing | To contribute your expertise and experience🙏💯 | Promptly🔥 | DLC Team |
| 🚧 GitHub DeepLabCut/Roadmap | To learn more about our journey |
N/A | N/A |
 |
To keep up with our latest news and updates 📢 | Daily | DLC Team |
| The DeepLabCut AI Residency Program | To come and work with us next summer👏 | Annually | DLC Team |
If you use this code or data we kindly ask that you please cite Mathis et al, 2018 and, if you use the Python package (DeepLabCut2.x) please also cite Nath, Mathis et al, 2019. If you utilize the MobileNetV2s or EfficientNets please cite Mathis, Biasi et al. 2021. If you use versions 2.2beta+ or 2.2rc1+, please cite Lauer et al. 2022.
DOIs (#ProTip, for helping you find citations for software, check out CiteAs.org!):
- Mathis et al 2018: 10.1038/s41593-018-0209-y
- Nath, Mathis et al 2019: 10.1038/s41596-019-0176-0
- Lauer et al 2022: 10.1038/s41592-022-01443-0
Please check out the following references for more details:
@article{Mathisetal2018,
title = {DeepLabCut: markerless pose estimation of user-defined body parts with deep learning},
author = {Alexander Mathis and Pranav Mamidanna and Kevin M. Cury and Taiga Abe and Venkatesh N. Murthy and Mackenzie W. Mathis and Matthias Bethge},
journal = {Nature Neuroscience},
year = {2018},
url = {https://www.nature.com/articles/s41593-018-0209-y}}
@article{NathMathisetal2019,
title = {Using DeepLabCut for 3D markerless pose estimation across species and behaviors},
author = {Nath*, Tanmay and Mathis*, Alexander and Chen, An Chi and Patel, Amir and Bethge, Matthias and Mathis, Mackenzie W},
journal = {Nature Protocols},
year = {2019},
url = {https://doi.org/10.1038/s41596-019-0176-0}}
@InProceedings{Mathis_2021_WACV,
author = {Mathis, Alexander and Biasi, Thomas and Schneider, Steffen and Yuksekgonul, Mert and Rogers, Byron and Bethge, Matthias and Mathis, Mackenzie W.},
title = {Pretraining Boosts Out-of-Domain Robustness for Pose Estimation},
booktitle = {Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV)},
month = {January},
year = {2021},
pages = {1859-1868}}
@article{Lauer2022MultianimalPE,
title={Multi-animal pose estimation, identification and tracking with DeepLabCut},
author={Jessy Lauer and Mu Zhou and Shaokai Ye and William Menegas and Steffen Schneider and Tanmay Nath and Mohammed Mostafizur Rahman and Valentina Di Santo and Daniel Soberanes and Guoping Feng and Venkatesh N. Murthy and George Lauder and Catherine Dulac and M. Mathis and Alexander Mathis},
journal={Nature Methods},
year={2022},
volume={19},
pages={496 - 504}}
@article{insafutdinov2016eccv,
title = {DeeperCut: A Deeper, Stronger, and Faster Multi-Person Pose Estimation Model},
author = {Eldar Insafutdinov and Leonid Pishchulin and Bjoern Andres and Mykhaylo Andriluka and Bernt Schiele},
booktitle = {ECCV'16},
url = {http://arxiv.org/abs/1605.03170}}
Review & Educational articles:
@article{Mathis2020DeepLT,
title={Deep learning tools for the measurement of animal behavior in neuroscience},
author={Mackenzie W. Mathis and Alexander Mathis},
journal={Current Opinion in Neurobiology},
year={2020},
volume={60},
pages={1-11}}
@article{Mathis2020Primer,
title={A Primer on Motion Capture with Deep Learning: Principles, Pitfalls, and Perspectives},
author={Alexander Mathis and Steffen Schneider and Jessy Lauer and Mackenzie W. Mathis},
journal={Neuron},
year={2020},
volume={108},
pages={44-65}}
Other open-access pre-prints related to our work on DeepLabCut:
@article{MathisWarren2018speed,
author = {Mathis, Alexander and Warren, Richard A.},
title = {On the inference speed and video-compression robustness of DeepLabCut},
year = {2018},
doi = {10.1101/457242},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2018/10/30/457242},
eprint = {https://www.biorxiv.org/content/early/2018/10/30/457242.full.pdf},
journal = {bioRxiv}}
This project is primarily licensed under the GNU Lesser General Public License v3.0. Note that the software is provided "as is", without warranty of any kind, express or implied. If you use the code or data, please cite us! Note, artwork (DeepLabCut logo) and images are copyrighted; please do not take or use these images without written permission.
SuperAnimal models are provided for research use only (non-commercial use).
- For all versions, please see here.
VERSION 3.0: A whole new experience with PyTorch🔥. While the high-level API remains the same, the backend and developer friendliness have greatly improved, along with performance gains!
VERSION 2.3: Model Zoo SuperAnimals, and a whole new GUI experience.
VERSION 2.2: Multi-animal pose estimation, identification, and tracking with DeepLabCut is supported (as well as single-animal projects).
VERSION 2.0-2.1: This is the Python package of DeepLabCut that was originally released in Oct 2018 with our Nature Protocols paper (preprint here). This package includes graphical user interfaces to label your data, and take you from data set creation to automatic behavioral analysis. It also introduces an active learning framework to efficiently use DeepLabCut on large experimental projects, and data augmentation tools that improve network performance, especially in challenging cases (see panel b).
VERSION 1.0: The initial, Nature Neuroscience version of DeepLabCut can be found in the history of git, or here: https://github.com/DeepLabCut/DeepLabCut/releases/tag/1.11
💜 We released a major update, moving from 2.x --> 3.x with the backend change to PyTorch
💜 The DeepLabCut Model Zoo launches SuperAnimals, see more here.
💜 DeepLabCut supports multi-animal pose estimation! maDLC is out of beta/rc mode and beta is deprecated, thanks to the testers out there for feedback! Your labeled data will be backwards compatible, but not all other steps. Please see the new 2.2+ releases for what's new & how to install it, please see our new paper, Lauer et al 2022, and the new docs on how to use it!
💜 We support multi-animal re-identification, see Lauer et al 2022.
💜 We have a real-time package available! http://DLClive.deeplabcut.org
-
June 2024: Our second DLC paper 'Using DeepLabCut for 3D markerless pose estimation across species and behaviors' in Nature Protocols has surpassed 1,000 Google Scholar citations!
-
May 2024: DeepLabCut was featured in Nature: 'DeepLabCut: the motion-tracking tool that went viral'
-
January 2024: Our original paper 'DeepLabCut: markerless pose estimation of user-defined body parts with deep learning' in Nature Neuroscience has surpassed 3,000 Google Scholar citations!
-
December 2023: DeepLabCut hit 600,000 downloads!
-
October 2023: DeepLabCut celebrates a milestone with 4,000 🌟 in Github!
-
July 2023: The user forum is very active with more than 1k questions and answers:
-
May 2023: The Model Zoo is now fully integrated into the DeepLabCut GUI, making it easier than ever to access a variety of pre-trained models. Check out the accompanying paper: SuperAnimal pretrained pose estimation models for behavioral analysis by Ye et al.
-
December 2022: DeepLabCut hits 450,000 downloads and 2.3 is the new stable release
-
August 2022: DeepLabCut hit 400,000 downloads
-
August 2021: 2.2 becomes the new stable release for DeepLabCut.
-
July 2021: Docs are now at https://deeplabcut.github.io/DeepLabCut and we now include TensorFlow 2 support!
-
May 2021: DeepLabCut hit 200,000 downloads! Also, Our preprint on 2.2, multi-animal DeepLabCut is released!
-
Jan 2021: Pretraining boosts out-of-domain robustness for pose estimation published in the IEEE Winter Conference on Applications of Computer Vision. We also added EfficientNet backbones to DeepLabCut, those are best trained with cosine decay (see paper). To use them, just pass "
efficientnet-b0" to "efficientnet-b6" when creating the trainingset! -
Dec 2020: We released a real-time package that allows for online pose estimation and real-time feedback. See DLClive.deeplabcut.org.
-
5/22 2020: We released 2.2beta5. This beta release has some of the features of DeepLabCut 2.2, whose major goal is to integrate multi-animal pose estimation to DeepLabCut.
-
Mar 2020: Inspired by suggestions we heard at this weeks CZI's Essential Open Source Software meeting in Berkeley, CA we updated our docs. Let us know what you think!
-
Feb 2020: Our review on animal pose estimation is published!
-
Nov 2019: DeepLabCut was recognized by the Chan Zuckerberg Initiative (CZI) with funding to support this project. Read more in the Harvard Gazette, on CZI's Essential Open Source Software for Science site and in their Medium post
-
Oct 2019: DLC 2.1 released with lots of updates. In particular, a Project Manager GUI, MobileNetsV2, and augmentation packages (Imgaug and Tensorpack). For detailed updates see releases
-
Sept 2019: We published two preprints. One showing that ImageNet pretraining contributes to robustness and a review on animal pose estimation. Check them out!
-
Jun 2019: DLC 2.0.7 released with lots of updates. For updates see releases
-
Feb 2019: DeepLabCut joined twitter
-
Jan 2019: We hosted workshops for DLC in Warsaw, Munich and Cambridge. The materials are available here
-
Nov 2018: We posted a detailed guide for DeepLabCut 2.0 on BioRxiv. It also contains a case study for 3D pose estimation in cheetahs.
-
Nov 2018: Various (post-hoc) analysis scripts contributed by users (and us) will be gathered at DLCutils. Feel free to contribute! In particular, there is a script guiding you through importing a project into the new data format for DLC 2.0
-
Oct 2018: new pre-print on the speed video-compression and robustness of DeepLabCut on BioRxiv
-
Sept 2018: Nature Lab Animal covers DeepLabCut: Behavior tracking cuts deep
-
Kunlin Wei & Konrad Kording write a very nice News & Views on our paper: Behavioral Tracking Gets Real
-
August 2018: Our preprint appeared in Nature Neuroscience
-
August 2018: NVIDIA AI Developer News: AI Enables Markerless Animal Tracking
-
July 2018: Ed Yong covered DeepLabCut and interviewed several users for the Atlantic.
-
April 2018: first DeepLabCut preprint on arXiv.org