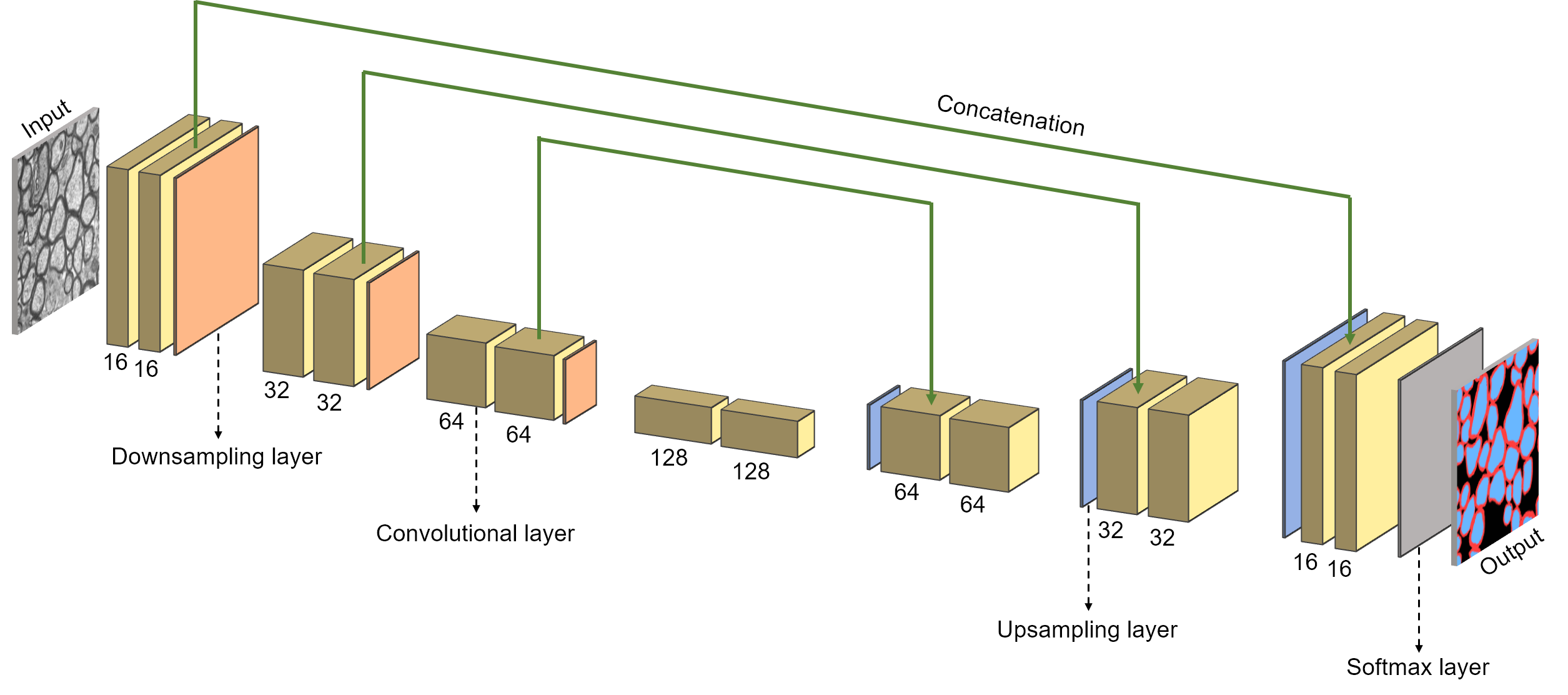
Segment axon and myelin from microscopy data using deep learning. Written in Python. Using the TensorFlow framework. Based on a convolutional neural network architecture. Pixels are classified as either axon, myelin or background.
For more information, see the documentation website.
Whether you are a newcomer or an experienced user, we will do our best to help and reply to you as soon as possible. Of course, please be considerate and respectful of all people participating in our community interactions.
- If you encounter difficulties during installation and/or while using AxonDeepSeg, or have general questions about the project, you can start a new discussion on the AxonDeepSeg GitHub Discussions forum. We also encourage you, once you've familiarized yourself with the software, to continue participating in the forum by helping answer future questions from fellow users!
- If you encounter bugs during installation and/or use of AxonDeepSeg, you can open a new issue ticket on the AxonDeepSeg GitHub issues webpage.
A tutorial demonstrating the basic features of our plugin for Napari is hosted on YouTube, and can be viewed by clicking this link.
AxonDeepSeg
- Lubrano et al. Deep Active Leaning for Myelin Segmentation on Histology Data. Montreal Artificial Intelligence and Neuroscience 2019 - [source code]
- Zaimi et al. AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks. Scientific Reports 2018
Applications
- Tabarin et al. Deep learning segmentation (AxonDeepSeg) to generate axonal-property map from ex vivo human optic chiasm using light microscopy. ISMRM 2019 - [source code]
- Lousada et al. Characterization of cortico-striatal myelination in the context of pathological Repetitive Behaviors. International Basal Ganglia Society (IBAGS) 2019
- Duval et al. Axons morphometry in the human spinal cord. NeuroImage 2019
- Yu et al. Model-informed machine learning for multi-component T2 relaxometry. Medical Image Analysis 2021 - [source code]
Reviews
If you use this work in your research, please cite it as follows:
Zaimi, A., Wabartha, M., Herman, V., Antonsanti, P.-L., Perone, C. S., & Cohen-Adad, J. (2018). AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks. Scientific Reports, 8(1), 3816. Link to paper: https://doi.org/10.1038/s41598-018-22181-4.
Copyright (c) 2018 NeuroPoly (Polytechnique Montreal)
The MIT License (MIT)
Copyright (c) 2018 NeuroPoly, École Polytechnique, Université de Montréal
Permission is hereby granted, free of charge, to any person obtaining a copy of this software and associated documentation files (the “Software”), to deal in the Software without restriction, including without limitation the rights to use, copy, modify, merge, publish, distribute, sublicense, and/or sell copies of the Software, and to permit persons to whom the Software is furnished to do so, subject to the following conditions:
The above copyright notice and this permission notice shall be included in all copies or substantial portions of the Software.
THE SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR OTHER DEALINGS IN THE SOFTWARE.
Pierre-Louis Antonsanti, Stoyan Asenov, Mathieu Boudreau, Oumayma Bounou, Marie-Hélène Bourget, Julien Cohen-Adad, Victor Herman, Melanie Lubrano, Antoine Moevus, Christian Perone, Vasudev Sharma, Thibault Tabarin, Maxime Wabartha, Aldo Zaimi.