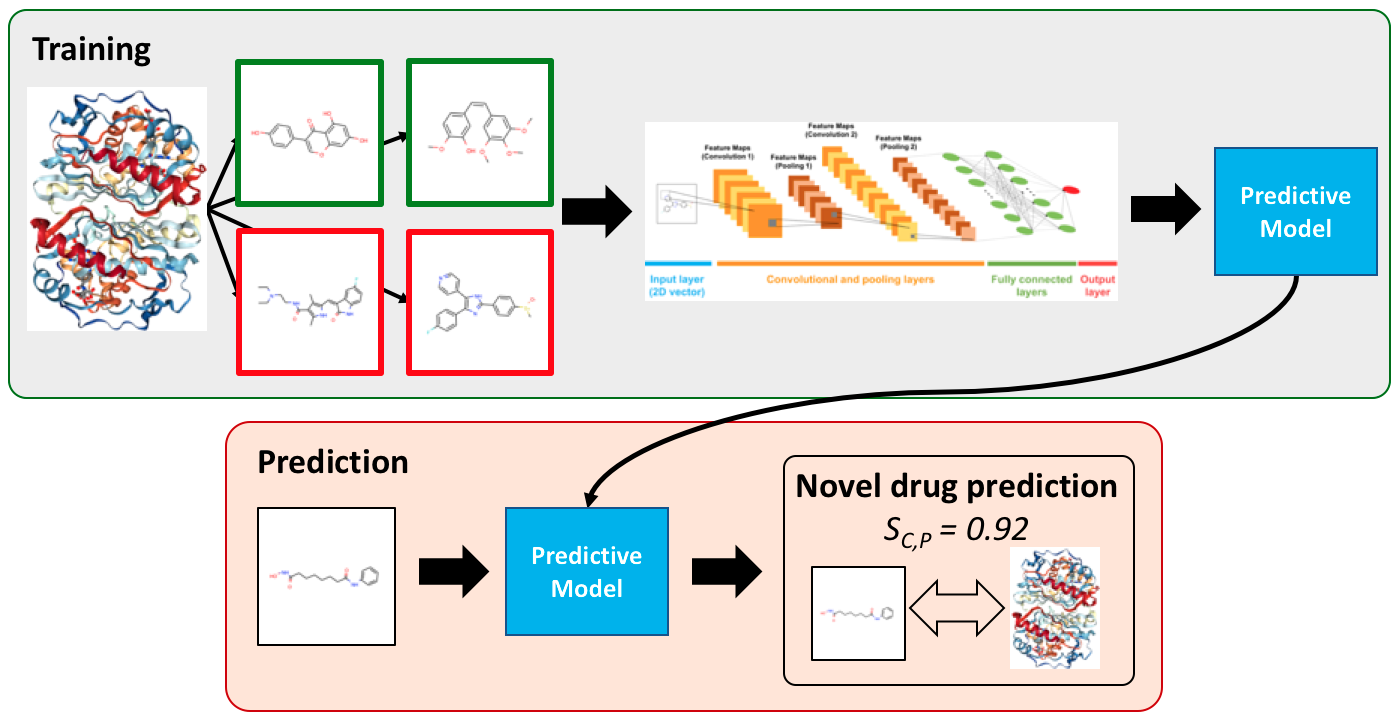
DEEPScreen is a large-scale DTI prediction system, for early stage drug discovery, using deep convolutional neural networks. One of the main advantages of DEEPScreen is employing readily available 2-D structural representations of compounds at the input level instead of conventional descriptors that display limited performance. DEEPScreen learns complex features inherently from the 2-D representations, thus producing highly accurate predictions. DEEPScreen system was trained for 704 target proteins (using curated bioactivity data) and finalized with rigorous hyper-parameter optimization tests.
-
bin This folder includes the source code of DEEPScreen
-
trainingFiles includes the files for training of the system.
- act_inact_comps_10.0_20.0_chembl_preprocessed_sp_b_pchembl_data.txt file contains active and inactive compound information before similarity-based negative training dataset enrichment for all targets. In this file, there are two lines for each target in the following format:
CHEMBL1790_act CHEMBL205013,CHEMBL96731,CHEMBL328791,... CHEMBL1790_inact CHEMBL306645,CHEMBL1765671,CHEMBL1765668,....The list of active/inactive compounds of the correnponding target separated by commas.
- act_inact_comps_10.0_20.0_chembl_preprocessed_sp_b_pchembl_data_blast_comp_20.txt file contains active and inactive compound information after similarity-based negative training dataset enrichment for all targets. The format of the file is same as above.
- DUDEDatasetFiles.zip contains training dataset for DUD-E dataset.
- Lenselink_Dataset_Files.zip contains training datasets for Lenselink et al.'s study.
- MUVDatasetFiles.zip contains training dataset for MUV dataset. and InChI representations for all ChEMBL compounds (version 23).
- chembl23_chemreps.txt.zip contains SMILEs and InChI representations for all ChEMBL compounds (version 23).
-
tempImage
- needed to create temporary images of compounds.
-
tflearnModels
- the folder that is used to store the trained models.
- trained-models can be dowloaded from here
-
resultFiles contains hyper-parameter values and performance calculations of optimized models.
- deepscreen_models_hyperparameters_performance_results.tsv stores hyper-parameter values and performance results of DEEPScreen models,
- dude_models_hyperparameters_performance_results.tsv stores hyper-parameter values and performance results of the final models trained with DUD-E dataset,
- lenselinks_models_hyperparameters_performance_results.tsv stores hyper-parameter values and performance results of the final models trained with Lenselink et al's dataset,
- muv_models_hyperparameters_performance_results.tsv stores hyper-parameter values and performance results of the final models trained MUV with dataset,
- Install dependencies and necessary libraries.
- Clone the DEEPScreen repository
- Decompress the zipped files
- Run DEEPScreen script by providing following command line arguments:
- DNN architecture (ImageNetInceptionV2, CNNModel)
- target ChEMBL ID
- optimizer type (adam, momentum, rmsprop)
- learning rate
- number of epochs
- number of neurons in the first fully-connected layer
- number of neurons in the second fully-connected layer
- drop-out keep rate
- save model (should be 1 to save the model or 0 (not save))
To train a model using the same hyper-parameters as DEEPScreen, you can use the hyper-parameters available in deepscreen_models_hyperparameters_performance_results.tsv which is located under resultFiles folder. Below is a sample command to train a model for target CHEMBL1790.
python trainDEEPScreen.py CNNModel CHEMBL1790 adam 0.0005 15 128 0 0.8 1
The evaluation results and predictions of independent test compounds are given as the output of this script. In the last line, the predictions for the test compounds are written in a tab-separated format where each field is separated by comma in the following format:
- <compound_id>,<prediction_outcome>,<true_label The ouput of the above command is written below as an example:
Test AUC:0.9445025083612041
Test AUPRC:0.9379908635681835
Test_f1score:0.88
Test_mcc:0.78
Test_accuracy:0.89
Test_precision:0.91
Test_recall:0.85
Test_tp:78
Test_fp:8
Test_tn:96
Test_fn:14
Test Predictions:
CHEMBL435331,TP,ACT CHEMBL3354592,TP,ACT CHEMBL44134,TN,INACT CHEMBL422701,TN,INACT CHEMBL105961,FN,ACT ...,
The name of the targets and hyper-parameter values are available in the following files
- dude_models_hyperparameters_performance_results.tsv,
- lenselinks_models_hyperparameters_performance_results.tsv,
- muv_models_hyperparameters_performance_results.tsv which are located under resultsFiles folder.
- To train DEEPScreen on MUV dataset
python trainConvNetMUV.py CNNModel MUV_692 adam 0.001 15 128 0 0.8 0
- To train DEEPScreen on DUD-E dataset
python trainDEEPScreenDUDE.py ImageNetInceptionV2 hdac8 adam 0.0001 5 0 0 0.8 0
- To train DEEPScreen on Lenselink et. al.'s dataset
python trainDEEPScreenLenselink.py ImageNetInceptionV2 CHEMBL274 adam 0.0001 5 0 0 0.8 0
The output of these commads same as the output shown above. Please note that you should unzip the corresponding folders (DUDEDatasetFiles.zip, MUVDatasetFiles.zip or Lenselink_Dataset_Files.zip) before running training scripts.
The model files should be located under tflearnModels folder. The model files for target CHEMBL1790 are put under tflearnModels folder as an example. Each target has a model which consists of three files. For our example, the the model files for or target CHEMBL1790 are as follows:
- CNNModel_CHEMBL1790_adam_0.0005_15_128_0.8_True-300.data-00000-of-00001
- CNNModel_CHEMBL1790_adam_0.0005_15_128_0.8_True-300.index
- CNNModel_CHEMBL1790_adam_0.0005_15_128_0.8_True-300.meta
Users should run loadDEEPScreenModel.py script to provide predictions for a set of compounds. The arguments of this script are as follows:
python loadDEEPScreenModel.py <target_name> <model_name> <path_to_smiles_of_compounds>
where <target_name> is the name of the ChEMBL target, <model_name> stands for the name of the model for the corresponding target stored under the tflearnModels folder and the last argument is the path to the SMILES of compounds. The SMILES file is a tab-seperated file with a header where the first column is compound identifier and the second colunmn is the smiles strings. You could have additional columns which are discarded by the script.There is a sample file (i.e. sample_test_compound_file.txt) under ../trainingFiles folder. You can run the following script to get the predictions for the compounds in the sample file.
python loadDEEPScreenModel.py CHEMBL1790 CNNModel_CHEMBL1790_adam_0.0005_15_128_0.8_True-300 ../trainingFiles/sample_test_compound_file.txt
The script provides the compund identifiers which are predicted as active as output.
ACTIVE PREDICTIONS:CHEMBL1790
CHEMBL350383
CHEMBL319636
CHEMBL182627
CHEMBL444956
CHEMBL331956
DEEPScreen Copyright (C) 2018 CanSyL
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see http://www.gnu.org/licenses/.