Shiitake mushrooms (Lentinula edodes) have over 200 cultivars and more than 300 IGS1 sequences available in INSD (the International Nucleotide Sequence Databases). This database integrates IGS1 sequences from shiitake strains across China, Japan, and Taiwan. It is designed to assist users in sequencing their shiitake strains and finding the closest matching sequences within the database, facilitating accurate identification and research.
Access the deployed version directly at GitHub Pages.
To use the database locally:
- Download this repository to your local computer.
- Open the
pairwise_alignment.htmlfile in your web browser.
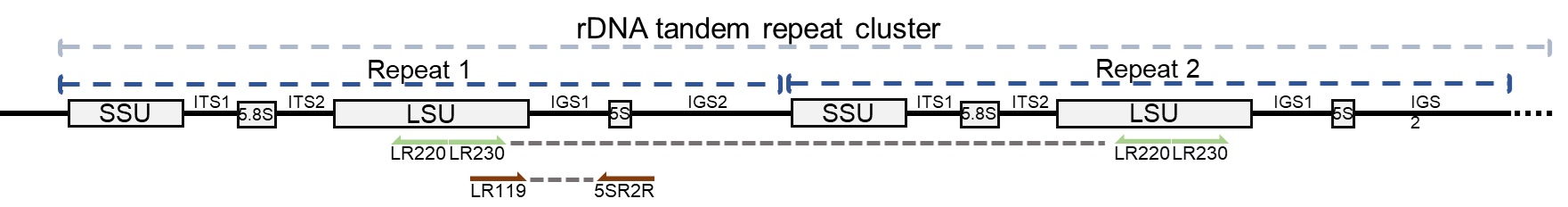
Shiitake DNA can be extracted using standard laboratory methods or commercial kits. Due to the high repeat number in the rDNA region where IGS1 is located, DNA can be successfully amplified from fresh or dried shiitake products, spawn, or spore powder.
1. Recommended Primers and PCR Program for IGS1 Region:
- Primers:
- LR119: GCGAAGCTATCATCTGCTGGA
- 5SR2R: TGGTCCCCCACCRTGGTACTAACT
- PCR Program:
- Initial denaturation: 94°C for 2 minutes
- 30-35 cycles of:
- 94°C for 30 seconds
- 60°C for 30 seconds
- 72°C for 60 seconds
- Final extension: 72°C for 60 seconds
2. Recommended Primers and PCR Program for Whole rDNA Region:
- Primers:
- LR230: CCACAGCCAAGGGGACGGGCTTGG
- LR220: CCCGCCGTTTACCCGCGCTTGG
- PCR Program:
- Initial denaturation: 98°C for 2 minutes
- 30-35 cycles of:
- 98°C for 20 seconds
- 68°C for 10 minutes
- Final extension: 68°C for 10 minutes
Note:
- If your shiitake strains have multiple IGS variants within the genome (as most do), you cannot directly send PCR samples for traditional Sanger sequencing. Instead, use a cloning kit to purify or consider using high-throughput sequencing.
- To amplify the full-length rDNA fragment (approximately 10 kb), it is recommended to use high-efficiency DNA polymerases with proofreading capability.
-
Upload Your Sequences:
- To upload the IGS1 sequences of your shiitake strains, use the provided input field. Sequences should be formatted in standard FASTA format. Each sequence must begin with a > symbol followed by the sequence title. Multiple sequences can be included by separating them with individual > symbols and titles.
-
Align Sequences:
- Click on the "Align Sequences" button to perform sequence alignment using the built-in Smith-Waterman algorithm.
-
View Results:
-
The results will be displayed in a table showing the most similar cultivars based on the uploaded sequences. The table includes strain names, similarity scores, and other relevant information. You can sort the results by clicking on the column headers.
-
Clicking on the strain name will display detailed alignment results, including the similarity matrix and alignment diagrams.
-
The detailed comparison window provides accession numbers, allowing users to link to the original sequences.
- If the user's strain matches a database strain, a typical similarity matrix will look like the one shown below. For example, Query Seq1 may match 22M0001 variant 2, and Query Seq2 may match 22M0001 variant 1.
-
-
Demo Mode:
- If you want to see a demonstration, click the "demo" button to load default query sequences (two IGS1 variants of MU30990, cultivar 588) and view the process and results.
This database is currently under review for publication. Once published, the citation details will be updated here. In the meantime, please cite this repository using its URL: https://github.com/Raingel/ShiitakeIGS1
The database is designed to accept sequence updates. In the root directory, there is a shiitake_list.csv file. Fill in the table with the required information, as shown in the example below, and submit a pull request. After review, the sequences will be added to the database and rebuilt using GitHub Actions.
Example:
| Cultivar | Strain | Isolation Source | Locality | Vendor | Accession 1 | Accession 2 | Accession 3 | Reference |
|---|---|---|---|---|---|---|---|---|
| Akiyama_A221 | Akiyama_A221 | commercial spawn | Japan | Akiyama Mycological Institute Co. Ltd. | AB251715 | AB251716 | (Babasaki et al., 2007) | |
| Akiyama_A526 | Akiyama_A526 | commercial spawn | Japan | Akiyama Mycological Institute Co. Ltd. | AB251721 | (Babasaki et al., 2007) | ||
| Akiyama_A567 | Akiyama_A567 | commercial spawn | Japan | Akiyama Mycological Institute Co. Ltd. | AB251722 | (Babasaki et al., 2007) | ||
| Kawamura_shokuyoukin_K5 | Kawamura_shokuyoukin_K5 | commercial spawn | Japan | Kawamura Shokuyoukin Kenkyujo Co. Ltd. | AB251765 | AB251804 | AB251805 | (Babasaki et al., 2007) |