Comments (23)
I have created a repository to evaluate a few popular tools.
@afni-dglen and @mrneont I am not very familiar with chauffeur_afni - as I provide the raw data, feel free to suggest better bitmaps that are more representative of this tool. Our goal would be to develop competitive features for NiiVue.
from niivue.
Hi, @neurolabusc -
Sure, thanks for the ping.
For specifically viewing edgy overlays, AFNI has a wrapper for @chauffeur_afni called @djunct_edgy_align_check, which is used extensively by afni_proc.py's automatic QC.
Running the following on your datasets:
@djunct_edgy_align_check \
-ulay example_func2highres.nii.gz \
-olay highres.nii.gz \
-box_focus_slices highres.nii.gz \
-use_olay_grid NN \
-prefix img_AEAC
... creates the following images:



Note that these AFNI-snapshotting programs produce 3 sets of images by default, one in each major viewing plane. The slices are evenly spread across each matrix grid, but one can also apply the "-box_focus_slices .." to trim the FOV usefully, as done here, so there aren't a lot of empty parts.
Note: these datasets are all oblique. I neither applied nor deleted their obliquity, but let the AFNI GUI deal with it (it looks fine here). I would maybe recommend testing with non-oblique data, which would cause regridding in some form if applying the obliquity.
--pt
from niivue.
Hi, @neurolabusc -
Thanks for updating the page on niivue with the AFNI tools.
However, I notice the AFNI-produced images on that other page:
https://github.com/neurolabusc/coreg_qa#afni
look different than what I posted here. The ones I ran, above, have a clear edge around the brain, but the ones on the linked page are missing this.
Looking at the code, it seems that one option was dropped out---the "-use_olay_grid NN"---which is necessary in this situation because the underlay dataset is an EPI, which sets the grid to be rather coarse. The find edges of the anatomical get lost in the resampling to the EPI without the above option.
Could that please be updated in the other main page?
thanks,
pt
from niivue.
@mrneont I think my local copy of AFNI is a bit out of date, as I do not have access to use_olay_grid. Its on the to do list to update things, but wanted a script I was sure @cdrake could emulate.
from niivue.
Also, I notice some images on the other page show the anatomical edges of the anatomical volume itself.
If it is useful to have an AFNI-produced comparison of that, you could include:
@djunct_edgy_align_check \
-ulay highres.nii.gz \
-olay highres.nii.gz \
-box_focus_slices highres.nii.gz \
-sharpen_ulay_off \
-prefix img_AEAC_a2a_reg
--pt
from niivue.
Re. #78 (comment)
Hmm, OK. That option is over a year old so I won't ask how old that version of AFNI binaries is...
From my point of view, having the full/modern version would be preferable, if possible, since it has been in use for a while. Or perhaps at least a note that users with modern AFNI should include that other option to see the full image (maybe showing one of those modern images)?
--pt
from niivue.
I did some experiments on this topic. I remain impressed with 3DEdge, also described here (though ftp links are non functional). It is unfortunate that it uses the GPL, so we can not adopt it. While I do think we can emulate the quality, the speed an low memory footprint of 3DEdge are really outstanding. In particular, the speed (which leverages the separable properties of the filter) is important for us, as JavaScript is pretty slow.
For my experiments, I have added a basic 3D Canny filter to niimath. The procedure is as described by Reeves. The procedure is:
- Remove outliers: Clamp values less than the robust range to the minimum value of the robust range. Clamp values greater than the robust range to the maximum value of the robust range.
- Apply a low-pass filter, a Gaussian blur with a sigma of 2 voxels seems suitable.
- Apply a Sobel filter that calculates the gradient magnitude, it also determines if the strongest gradient direction is along the rows, columns or slices.
- Normalize the gradient magnitude to the range 0..1. Set any values below 0.1 to zero (lower bound cut-off suppression).
- Edge tracking: to survive, an edge must be have a stronger gradient in its direction (row, column, slice) than its neighbor. In other words, if this is a
rowedge (gX was strongest gradient), it must have a stronger gradient magnitude than its neighbor to the left or right (this makes the edge a single voxel skeleton). Likewise, to survive it must have some neighbors that are also non-zero gradients (e.g. arowedge must have a non-zero magnitude neighbor along thecolumnorslicedimension). This removes unconnected noise.
With the latest commit to niimath, this can be applied using the call:
./niimath highres.nii.gz -clamp 0 -uclamp 100 -sv 2 -sobel_binary outv
Like AFNI and FSL, this creates a binarized image. One nice idea I saw was the work by Bahnisch et al., 2009 that provides sub-voxel edges to capture partial volume effects. It is a shame they do not provide an implementation of their procedure. They also omit the edge tracing step, simply saying it is complicated. Regardless, perhaps something to pursue. A very simple alternative is to just add a little blur to the skeleton:
./niimath highres.nii.gz -clamp 0 -uclamp 98 -sv 2 -sobel_binary -sv 1 -range outv
from niivue.
Hi, @neurolabusc -
That is looking good. Glad 3dedge3 has been a useful starting point.
I would just note that 3dedge3 does not create a binarized edge map: steeper edges have higher values than shallow ones.
In the above images from the overlap_check wrapper, this is particularly nice with a yellow-to-red colorbar, because shallower edges are fainter (but still present) while "major" edges pop out visually.
--pt
from niivue.
The Difference of Gassian (DoG) (which approximates the Laplacian of Gaussian) provides another elegant solution. I added an implementation that colors zero crossings. The voxel gets a color of 1 if there is a zero crossing at the voxel face, 0.5 if it is at the voxel edge, and 0.25 if the crossing is at a corner. This creates nice, smooth boundaries. This implementation creates two voxel thick borders.
./niimath example_func2highres.nii.gz -clamp 10 -uclamp 100 -dog_edge 2 4 edge
from niivue.
The latest commit allows single or double voxel cases. In both cases, a difference of Gaussian is calculated. If the user specifies dog1 the edge is defined as positive voxels that share negative voxel neighbors. If dog2 is specified, both positive and negative voxels with zero-crossing neighbors are counted as edges. So calling might look like this:
niimath example_func2highres -clamp 0 -uclamp 100 -dog1 2 4 epi1
niimath example_func2highres -clamp 0 -uclamp 100 -dog2 2 4 epi2
niimath highres -clamp 0 -uclamp 100 -dog1 2 4 struct1
niimath highres -clamp 0 -uclamp 100 -dog2 2 4 struct2
In these cases, the single voxel edge is on the bright side of the boundary. However, you can reverse the order of the positive and negative Gaussian to define the edge as the dark side:
niimath highres -clamp 0 -uclamp 100 -dog1 4 2 struct1
I find these edges very visually pleasing. The Gaussian function is separable, so it is pretty computationally fast. However, it does use a lot of memory (float copies of both blurred images must be stored). For contemporary computers and MRI resolutions, the demands are pretty modest (3DEdge3 was written in 1998).
from niivue.
Assirati et al. suggest one can use Q-Gaussian to derive a better approximation of the Laplacian of Gaussians. However, the classic Gaussian has a nice property of being separable, and the tiny amount of blurring is probably advantageous for MRI scans with modest signal to noise.
from niivue.
Those images look quite nice.
from niivue.
Another nice trick for thin lines is to create the 2-voxel dog and then apply an unsharp mask. I have updated the dog function so the filter can be specified in voxels using negative values (-dog -2 -4 uses a Sigma of 2 and 4 voxels) with positive values signifying mm (-dog 2 4 for Sigma 2 and 4mm). My sense is that mm makes sense, since the features of interest have a known size (e.g. cortex is ~2.5mm thick).
niimath highres -clamp 0 -uclamp 100 -dog1 1.5 3 -unsharp 1 1 st1
Maybe @jonclayden has thoughts as his imbibe is a wrapper for niimath and his mmand has functions for relevant features like skeletonisation.
I do feel that the Difference of Gaussian is a nicer solution for brain imaging than the Canny-like edge detectors. The brain has features of known physical size in mm, so the DoG parameters seem like constants. With the Canny-style edge filters, one must select some arbitrary edge threshold (I concede 3dedge3 seems to have a more robust heuristic than FSL's slicer or my own attempts).
from niivue.
I must admit I haven't put much thought into this – certainly I haven't done anything like as much experimentation as @neurolabusc – so I don't think I can add anything very helpful to this informative thread.
TractoR's reg-check script, which aims to serve this purpose, just calculates a morphological gradient using a basic kernel and then uses k-means to threshold the result. It's an unsophisticated approach that I committed fairly hastily back in 2016 and essentially haven't revisited. On this example it gives a reasonable, if rather thick, outline, but doesn't do very well with internal details.
from niivue.
@jonclayden thanks. The dilation/erosion combination is an interesting trick, and looks pretty similar to the simple Sobel both in performance and computational cost. As you note, these miss internal structures.
from niivue.
Yes, indeed. I'll have to try out some of the alternatives you've mentioned here at some point. Thanks for posting the results of so many interesting experiments!
from niivue.
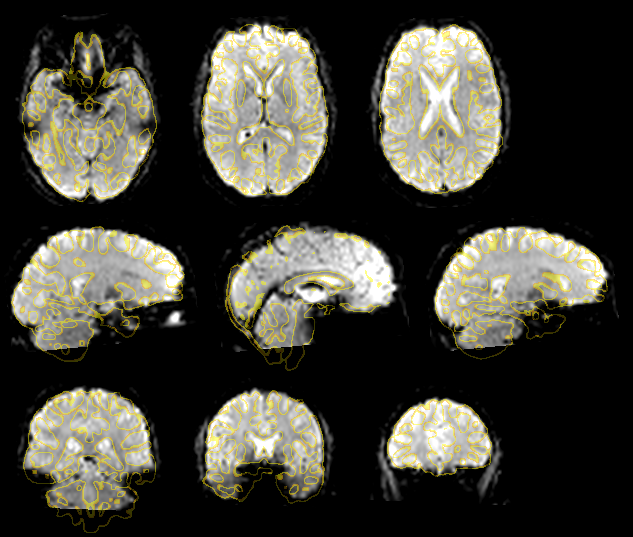
Interesting discussion. We found "edginess" is really useful for checking alignment. chauffeur_afni and @AddEdge use 3dedge3 for a nice edge view.
The AFNI GUI also has the Edge Enhance button (either tap 'e' on the image viewer, or select in the Display panel). That uses a Sobel filter described in mri_sobel.c in the AFNI source code. It's applied in 2D on that particular slice view.
I tried some quick alternatives. From left to right in the image below:
A. 3dedge3 -Deriche Sobel filter implementation
B. Spatial variance. Average difference in 27 voxel neighborhood normalized by mean (stdev/fabs(mean)), thresholded at around 50%ile.
C. Flux image. L2 norm of second difference in x,y,z directions computed as part of 3D anisotropic smoothing, thresholding at around 63%ile.
D. Spatial variance of spatial variance (C). Poor man's second derivative. 29%ile thresholding.
E. Edge of Spatial variance^2 (D), preserves only the edge by one voxel simple erosion to find outer periphery (corners, edges, faces - all treated equivalently).
The color scales are not the same across all these, and lots of tricks can be played to manipulate the colors to enhance edges. Still I haven't done too much here. I think 3dedge3 still provides the best of these, and without fiddling around for specific thresholds.
from niivue.
Did some work to refine the DoG implementation:
- I refactored the Gaussian smooth code in niimath that will make it easier to port to JavaScript. It could be simplified further if we only want to use a single thread, or if we use multiple threads only to process 4D datasets.
- I realized there is a simple performance optimization for DoG: this method operates by creating two smoothed images: one with a narrow width kernel (e.g. 2.5mm FWHM) and one with a larger width (e.g. 5mm FWHM). Since the operations for a separable 3D Gaussian are
O(xyzi)+O(xyzj)+O(xyzk)(where xyz are dim[1..3] and ijk are the filter kernel widths), the wider kernel is necessarily slower than the smaller one. However, we can use the results of the narrow kernel as the input for the wider kernel, and reduce its width (and therefore increase speed). In other words, doing a 2.5mm FWHM blur twice results in the same amount of blur as doing a 3.53mm blur. The results of the narrow kernel are free, as we need to compute it anyways. - In practice, the performance of a 3D Gaussian blur is often constrained by the transposing of data. In other words, we have hit the memory wall. One trick is to transpose tiles to use coherent cache reads, further (though probably not useful for JavaScript) there are some SIMD instructions to accelerate this like _MM_TRANSPOSE4_PS. For 3D operations we need to transpose both within slices (TransposeXY) and across slices (TransposeXZ). The latter has less cache coherency, and in my testing with a 256^3 voxel volume takes about four times longer.
from niivue.
I notice that bisweb has a WASM 3D Gaussian we can use (thanks to their Apache license). Xenios (@bioimagesuiteweb) is a consultant for NiiVue. Our tools fit different niches, and we should leverage their well documented and tested solutions wherever possible.
from niivue.
We also have a pure JS implementation if you prefer that -- this predates the C++/WASM version.
from niivue.
https://github.com/bioimagesuiteweb/bisweb/blob/devel/js/utilities/bis_imagesmoothreslice.js
from niivue.
this will be part of niimath wasm
from niivue.
@XeniosP and @hanayik niimath is now ported to WASM. In general, it is a superset of fslmaths. The 3D Gaussian uses optimizations not found in bisweb, and the additional functions may make this interesting for bisweb.
You can evaluate this WASM implementation from the command line using the niimath.js project, which is a rough clone of fslmaths:
git clone --branch development https://github.com/rordenlab/niimath
cd niimath/src
emcc -O2 -s ALLOW_MEMORY_GROWTH -s MAXIMUM_MEMORY=4GB -s TOTAL_MEMORY=268435456 -s WASM=1 -DUSING_WASM -I. core32.c nifti2_wasm.c core.c walloc.c -o funcx.js
node test.js
npm install nifti-reader-js
node niimath.js ~/T1.nii.gz -dehaze 5 -dog 2 3.2 ~/dT1.nii
from niivue.
Related Issues (20)
- Dependencies not included in saved HTML script string
- TRK format implementations
- Crosshair gap HOT 1
- Restore legendLineThickness
- setMultiplanarLayout regression
- Plot layout issues
- Retore outline shaders
- AFNI volumes animation HOT 1
- Multiuser colormaps live demo regression
- Multiuser basic 3D live demo regression
- Connectome refactor and consistency
- Remove Websocket comms from niivue
- Support html scene export via downloadable "big" build.
- remove puppeteer
- Labelmap intensity range not set correctly when cal_min and cal_max are incorrect HOT 5
- clone() should assign a new ID rather than using the donor image ID
- Dicom Segmentation not supported HOT 4
- Clusterize volumes
- Self-generate volumes
- Conform volumes
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.












from niivue.