Comments (4)
Hello,
Thanks for trying out Numbat. Could you attach the bulk_subtrees_1.png as well?
In general, there are three scenarios that a CNV is not called. I'm thinking of making a FAQ about this since many people have asked.
-
The CNV is not called in pseudobulk HMM. You can check bulk_subtrees_*.png to see if the event was called. If it was not, I would suggest trying to find a better expression reference to reduce the noise in logFC, or changing the parameters used to configure the HMM (
gamma,t, etc). Sometimes the CNV is very subclonal and was not isolated as a pseudobulk by the initial clustering. You can see if this was the case by looking at the exp_roll_clust.png and if so, try increasing k_init. -
CNV is called in pseudobulks but did not appear in
segs_consensus_*.tsvandjoint_post_*tsv. This is because the event is filtered out due to low LLR. Loweringmin_LLRthreshold will help. -
CNV is called in pseudobulks, is present in
segs_consensus_*.tsvandjoint_post_*tsv, but did not appear in phylogeny. This is because the event is filtered out due to high entropy (weak evidence) in single cells. Raisingmax_entropythreshold will help. You can check the entropy of specific events injoint_post_*tsvin the columnavg_entropy.
Best,
Teng
from numbat.
Thanks @teng-gao, for the detailed answer! Below the bulk_subtrees_1.png
I will give a try to the several recommendations/options you have given!
It is a wonderful tool, and I am really happy with the performance! And kudos for the publication!
from numbat.
Thanks @ccruizm! Based on the pseudobulk profiles, the allelic frequency is quite balanced on all chromosomes so I suspect there aren't actually CNVs there (does WES show allelic imbalance?). The only other possibility is that all CNVs are balanced gains and losses, which is extremely rare.
from numbat.
I am waiting for the WES report. In the meantime, I checked the exp_roll_clust.png, which looks pretty similar to the one from copyKAT.
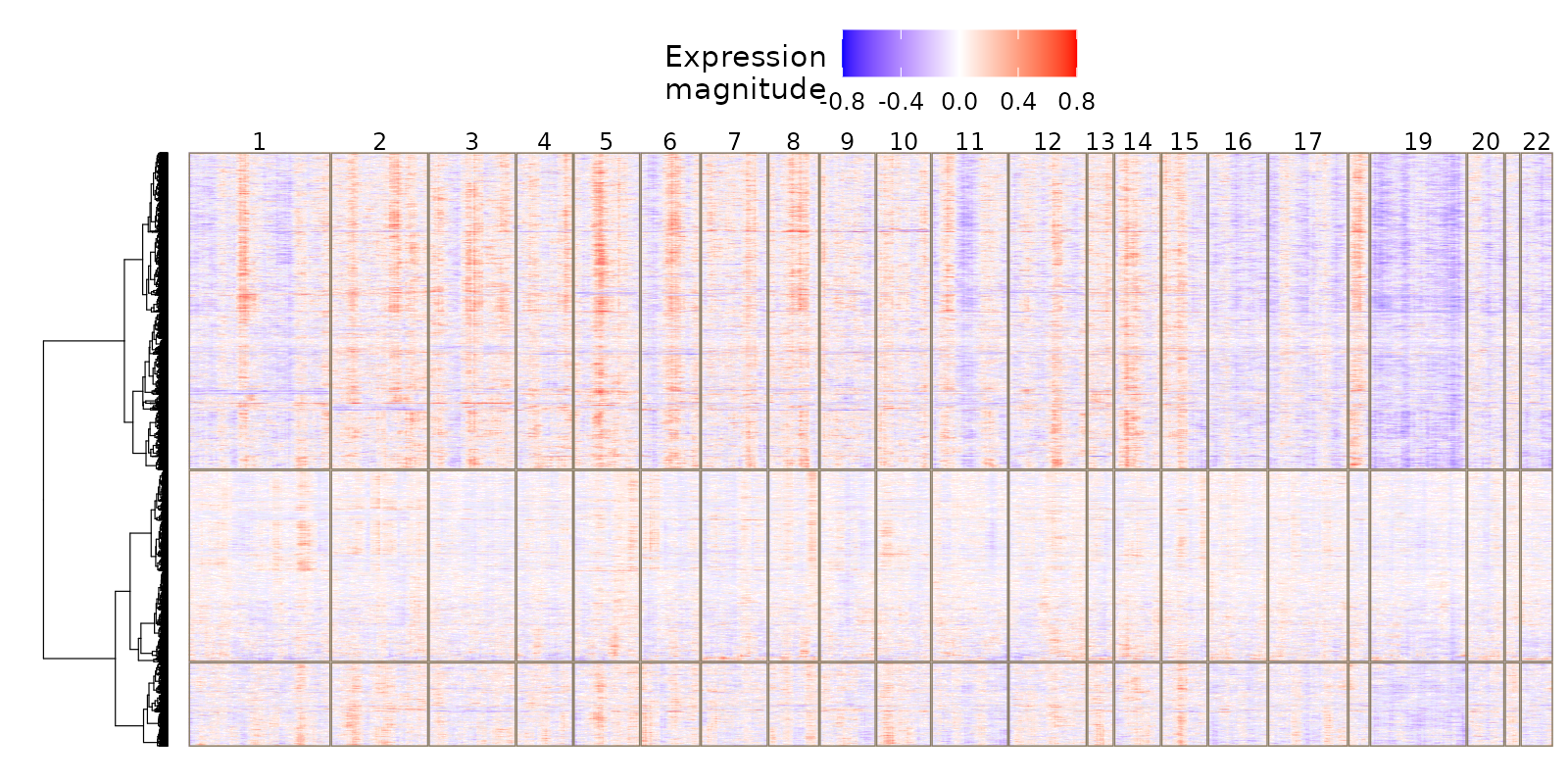
I increased the k_init (10) and now it detected some iCNV but actually they seems to be inverted of what the expression heatmaps show.
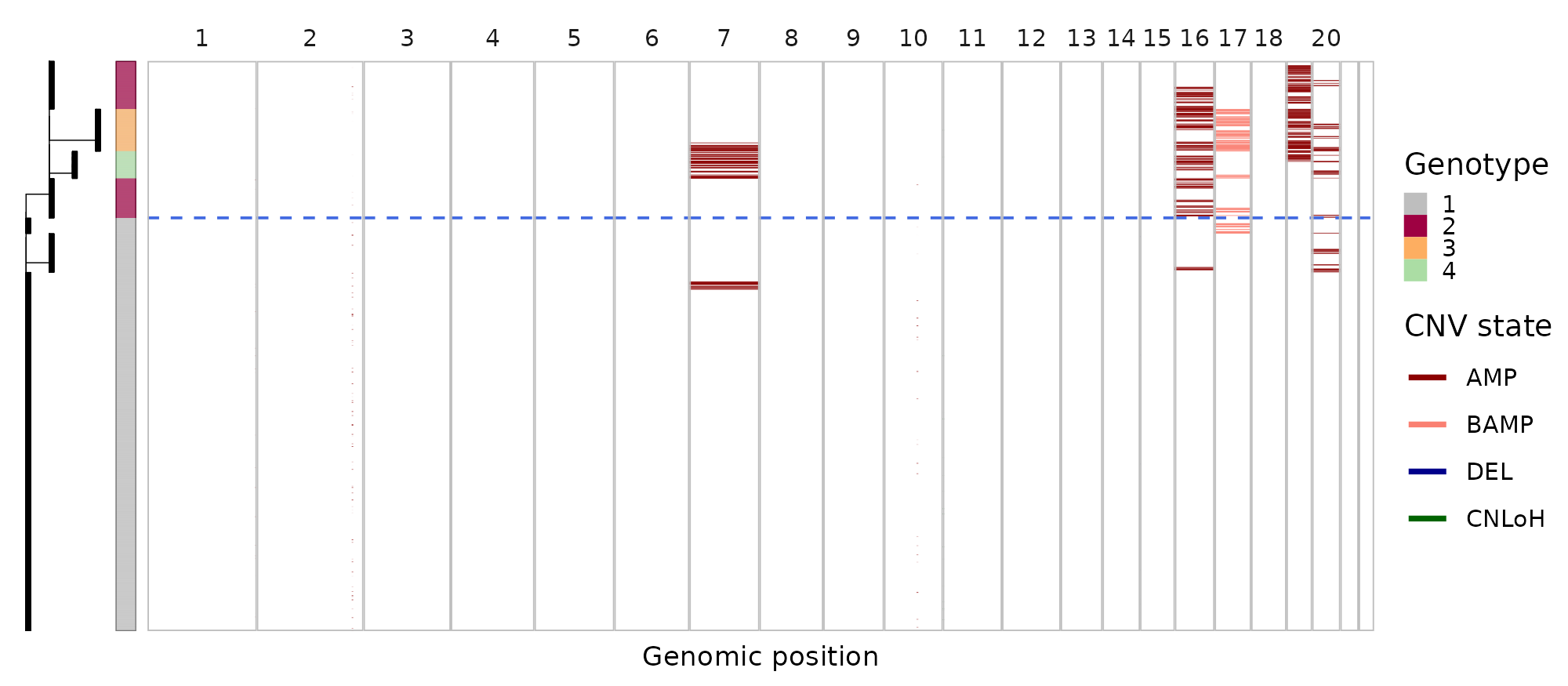
What do you think might be the reason of this incongruency? Thanks again for your help!
from numbat.
Related Issues (20)
- memory usage error during run_numbat
- why genotype1 have tumor cells? HOT 1
- High Similarity of Clones' Profiles
- How to use numbat in other species with a phased genome? HOT 3
- Chromosome Arms Summaries File Lacking
- Running numbat in multiple samples from same patient HOT 1
- Error in mutate(., state = run_allele_hmm ... Not a matrix HOT 2
- n_states is 0 for CNV region with multiallelic CNVs are not called
- Better error handling when one of the pileup fails
- No SNPs left HOT 1
- ```run_numbat()``` error unclear HOT 4
- bulk_clones plot "CNV state" legend colors HOT 2
- Excluding segs_loh causes multiple CNV states for the same segment HOT 2
- Duplicated segments in segs_consensus for NCI-N87
- saving numbat object after changing n_cut=k kvalue
- Fail during numbat : object 'seg' not found
- Input only the gene expression matrix HOT 2
- segfault, caught bus error, non-existent physical address HOT 6
- Error because of dplyr version
- Error: 'separate_longer_delim' is not an exported object from 'namespace:tidyr' HOT 1
Recommend Projects
-
 React
React
A declarative, efficient, and flexible JavaScript library for building user interfaces.
-
Vue.js
🖖 Vue.js is a progressive, incrementally-adoptable JavaScript framework for building UI on the web.
-
 Typescript
Typescript
TypeScript is a superset of JavaScript that compiles to clean JavaScript output.
-
TensorFlow
An Open Source Machine Learning Framework for Everyone
-
Django
The Web framework for perfectionists with deadlines.
-
Laravel
A PHP framework for web artisans
-
D3
Bring data to life with SVG, Canvas and HTML. 📊📈🎉
-
Recommend Topics
-
javascript
JavaScript (JS) is a lightweight interpreted programming language with first-class functions.
-
web
Some thing interesting about web. New door for the world.
-
server
A server is a program made to process requests and deliver data to clients.
-
Machine learning
Machine learning is a way of modeling and interpreting data that allows a piece of software to respond intelligently.
-
Visualization
Some thing interesting about visualization, use data art
-
Game
Some thing interesting about game, make everyone happy.
Recommend Org
-
Facebook
We are working to build community through open source technology. NB: members must have two-factor auth.
-
Microsoft
Open source projects and samples from Microsoft.
-
Google
Google ❤️ Open Source for everyone.
-
Alibaba
Alibaba Open Source for everyone
-
D3
Data-Driven Documents codes.
-
Tencent
China tencent open source team.


from numbat.